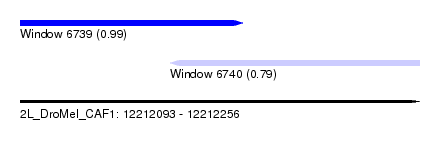
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,212,093 – 12,212,256 |
| Length | 163 |
| Max. P | 0.992308 |

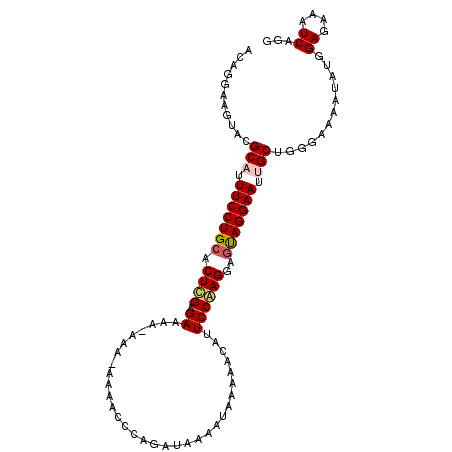
| Location | 12,212,093 – 12,212,184 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 91.84 |
| Mean single sequence MFE | -12.93 |
| Consensus MFE | -11.50 |
| Energy contribution | -11.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992308 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12212093 91 + 22407834 UCGUCGCGACAUUUCUACUGUGUCUCGACUGCGUUUCCCUU-UUCCCCAGA-UUCACCAGAUCCCUGAUUUCUCCAUAUUUUCCCAGCGAUUC ..((((.(((((.......))))).))))..((((......-......(((-.....(((....)))...)))............)))).... ( -14.75) >DroSec_CAF1 40802 92 + 1 UCGUCGCGACAUUUCUACUGUGUCUCGACUGCGUUUCCCUU-UUCCCCAGACUUCCCCAGAUCCCAGAUUUCUCCAUAUUUUCCCAGCCAUUC ..((((.(((((.......))))).)))).((((((.....-......))))......(((((...)))))...............))..... ( -11.70) >DroSim_CAF1 39497 92 + 1 UCGUCGCGACAUUUCUACUGUGUCUCGACUGCAUUUCCCUU-UUCCCCAGACUUCCCCAGAUCCCAGAUUUCUCCAUAUUUUCCCAGCAAUUC ..((((.(((((.......))))).))))(((.........-.((....)).......(((((...)))))...............))).... ( -12.30) >DroEre_CAF1 39118 87 + 1 UCGUCGCGACAUUUCUACUGUGUCUCGACUGCGUUUCCCUU-UUCCCCAGAU-UCC----AUCCCUGAUUUCUCCAUAUUUUCCCGGCAAUUC ..((((.(((((.......))))).))))............-.....(((..-...----....))).......................... ( -12.20) >DroYak_CAF1 39788 88 + 1 UCGUCGCGACAUUUCUACUGUGUCUCGACUGCGUUUCCCUUUUUCCCCAGAU-UCC----AUUCCUGAUUUCUCCAUGUUUUCCCAGCAAUUC ..((((.(((((.......))))).))))(((...............(((..-...----....))).........((......))))).... ( -13.70) >consensus UCGUCGCGACAUUUCUACUGUGUCUCGACUGCGUUUCCCUU_UUCCCCAGACUUCCCCAGAUCCCUGAUUUCUCCAUAUUUUCCCAGCAAUUC ..((((.(((((.......))))).))))................................................................ (-11.50 = -11.50 + 0.00)

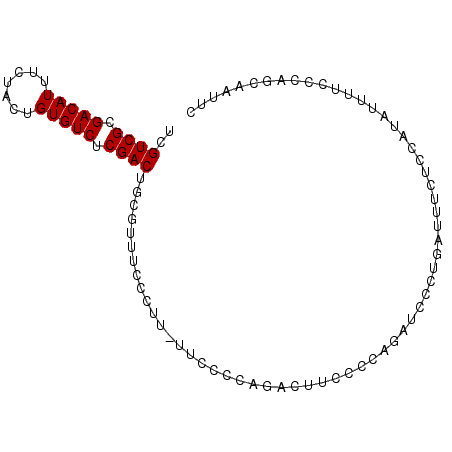
| Location | 12,212,154 – 12,212,256 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 91.45 |
| Mean single sequence MFE | -21.13 |
| Consensus MFE | -15.49 |
| Energy contribution | -16.05 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.790446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12212154 102 - 22407834 ACAGGAAGUACGCAUUUCCUGCACUUGAGAAAA-AAA-AAAACACAGAUAAAAUAAAAACAUUCCAAGGAGUAGGAAUCGCUGGGAAAAUAUGGAGAAAUCAGG ...........((..(((((((.((((.(((..-...-.......................)))))))..)))))))..))............((....))... ( -17.19) >DroSec_CAF1 40864 103 - 1 ACAGGAAGUACGCAUUUCCUGAACUCGAGAUAAAAAA-AAAACCCAGAUAAAAUAAAAACAUUCCGAGGAGUAGGAAUGGCUGGGAAAAUAUGGAGAAAUCUGG .((((((((....))))))))................-....(((((............((((((........)))))).))))).......(((....))).. ( -26.72) >DroSim_CAF1 39559 104 - 1 ACAGGAAGUACGCAUUUCCUGCACUCGAGAUAAAAAAAAAAAACCAGAUAAAAUAAAAACAUUCCGAGGAGUAGGAAUUGCUGGGAAAAUAUGGAGAAAUCUGG .((((((((....))))))))......................((((((............((((.((.(((....))).)).))))...........)))))) ( -22.20) >DroYak_CAF1 39846 99 - 1 ACAGGAAGUACGCAUUUCCUGCACUCGAGAAAA-----AAAACGCAGAUAAAAUAAAAACAUUCCAAGGAGCAGGAAUUGCUGGGAAAACAUGGAGAAAUCAGG ...........(((.(((((((.((((......-----....)).))...............((....))))))))).)))............((....))... ( -18.40) >consensus ACAGGAAGUACGCAUUUCCUGCACUCGAGAAAA_AAA_AAAACCCAGAUAAAAUAAAAACAUUCCAAGGAGUAGGAAUUGCUGGGAAAAUAUGGAGAAAUCAGG ...........(((.(((((((.((((.((................................))))))..))))))).)))............((....))... (-15.49 = -16.05 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:55 2006