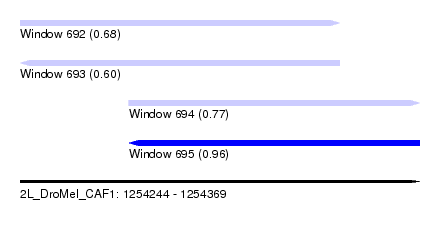
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,254,244 – 1,254,369 |
| Length | 125 |
| Max. P | 0.963250 |

| Location | 1,254,244 – 1,254,344 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 76.72 |
| Mean single sequence MFE | -21.24 |
| Consensus MFE | -11.20 |
| Energy contribution | -11.96 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679582 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1254244 100 + 22407834 AAUGCGAA--AAGUGCCUUCAACUUUUUAUGAAAUAUGGGUUGACAGACGGCUCAUCUGUCAUCUAAUAAAUGCAUUUAAAAUUAAACAAAUGCGCGCAUA-------A .(((((((--((((.......)))))))........(((((.((((((.......)))))))))))......((((((..........))))))..)))).-------. ( -22.60) >DroSec_CAF1 13969 100 + 1 AAUGCGAA--AAGUGCUCUCUACUUUUUAUGAAAUAUGGGUUGACAGAUGGCUCAUCUGUCAUCUAAUAAAUGCAUUUAAAAUUAAACAAAUGCGCGAAGA-------A ..((((((--(((((.....))))))))........(((((.((((((((...)))))))))))))......((((((..........)))))))))....-------. ( -24.30) >DroSim_CAF1 14471 100 + 1 AAUGCGAA--AAGUGCUUCCUACUUUUUAUGAAAUAUGGGAUGACAGAUGGCUCAUCUGUCAACUAAGAAAUGCAUUUAAAAUUAAACAAAUGCGCGCAAA-------A ..((((..--.(((...(((((..(((....)))..)))))(((((((((...)))))))))))).......((((((..........)))))).))))..-------. ( -25.60) >DroEre_CAF1 26783 81 + 1 AAUGCGGAGGAAGUGCUCCCUACUUUUUAUG------------ACAGACGGCUCAUCUGCCAUCCAAUAAACGCGUUUCGAACUAAA----UU-----AAA-------A ((((((.((((((((.....))))))))...------------...((.(((......))).)).......))))))..........----..-----...-------. ( -15.70) >DroYak_CAF1 14628 102 + 1 AAUGCGGA--AAGUGCUUCCUACUUUUUAUGAAAUAUGGGUUGACAGGCGGCUCAUCUGUCAUCUAAGAAACGCGAUUGAAAUUAAACGAAUU-----AAAAUAUGAAG ..((((((--(((((.....))))))).((((...((((((((.....))))))))...))))........))))..................-----........... ( -18.00) >consensus AAUGCGAA__AAGUGCUUCCUACUUUUUAUGAAAUAUGGGUUGACAGACGGCUCAUCUGUCAUCUAAUAAAUGCAUUUAAAAUUAAACAAAUGCGCG_AAA_______A ((((((....(((((.....))))).............((.(((((((.......))))))).))......))))))................................ (-11.20 = -11.96 + 0.76)

| Location | 1,254,244 – 1,254,344 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 76.72 |
| Mean single sequence MFE | -21.26 |
| Consensus MFE | -11.92 |
| Energy contribution | -12.00 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1254244 100 - 22407834 U-------UAUGCGCGCAUUUGUUUAAUUUUAAAUGCAUUUAUUAGAUGACAGAUGAGCCGUCUGUCAACCCAUAUUUCAUAAAAAGUUGAAGGCACUU--UUCGCAUU .-------.(((((.(((((((........)))))))..........(((((((((...))))))))).......(((((........)))))......--..))))). ( -24.30) >DroSec_CAF1 13969 100 - 1 U-------UCUUCGCGCAUUUGUUUAAUUUUAAAUGCAUUUAUUAGAUGACAGAUGAGCCAUCUGUCAACCCAUAUUUCAUAAAAAGUAGAGAGCACUU--UUCGCAUU .-------((((..((((((((........)))))))..........(((((((((...)))))))))..................)..))))((....--...))... ( -24.30) >DroSim_CAF1 14471 100 - 1 U-------UUUGCGCGCAUUUGUUUAAUUUUAAAUGCAUUUCUUAGUUGACAGAUGAGCCAUCUGUCAUCCCAUAUUUCAUAAAAAGUAGGAAGCACUU--UUCGCAUU .-------..((((.(((((((........)))))))........(((((((((((...))))))))...((.(((((......))))))).)))....--..)))).. ( -24.30) >DroEre_CAF1 26783 81 - 1 U-------UUU-----AA----UUUAGUUCGAAACGCGUUUAUUGGAUGGCAGAUGAGCCGUCUGU------------CAUAAAAAGUAGGGAGCACUUCCUCCGCAUU .-------...-----..----.............((..(((((..((((((((((...)))))))------------)))....)))))((((......))))))... ( -20.70) >DroYak_CAF1 14628 102 - 1 CUUCAUAUUUU-----AAUUCGUUUAAUUUCAAUCGCGUUUCUUAGAUGACAGAUGAGCCGCCUGUCAACCCAUAUUUCAUAAAAAGUAGGAAGCACUU--UCCGCAUU ...........-----...................(((.........((((((.........))))))..((.(((((......)))))))........--..)))... ( -12.70) >consensus U_______UUU_CGCGCAUUUGUUUAAUUUUAAAUGCAUUUAUUAGAUGACAGAUGAGCCGUCUGUCAACCCAUAUUUCAUAAAAAGUAGGAAGCACUU__UUCGCAUU ................................(((((..........(((((((((...))))))))).....................(((.........)))))))) (-11.92 = -12.00 + 0.08)

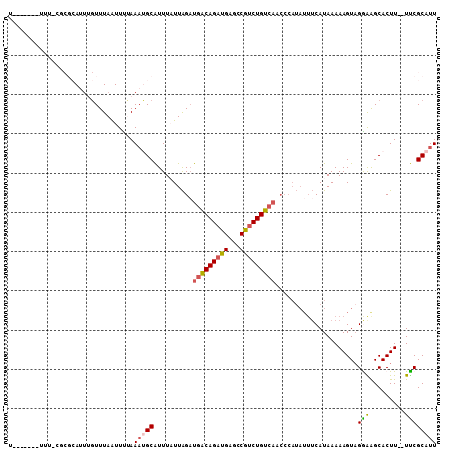
| Location | 1,254,278 – 1,254,369 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 91.58 |
| Mean single sequence MFE | -22.33 |
| Consensus MFE | -18.27 |
| Energy contribution | -18.60 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1254278 91 + 22407834 UGGGUUGACAGACGGCUCAUCUGUCAUCUAAUAAAUGCAUUUAAAAUUAAACAAAUGCGCGCAUAAGUGCUGCAAUCAGUUAGACAUUAUG .....(((((((.......)))))))((((((....((((((..........))))))((((....))))........))))))....... ( -20.70) >DroSec_CAF1 14003 91 + 1 UGGGUUGACAGAUGGCUCAUCUGUCAUCUAAUAAAUGCAUUUAAAAUUAAACAAAUGCGCGAAGAAGUGCUGCAAUAAGUUAGAUACCAUG (((...((((((((...))))))))(((((((...((((((((....)))).....((((......))))))))....))))))).))).. ( -25.30) >DroSim_CAF1 14505 91 + 1 UGGGAUGACAGAUGGCUCAUCUGUCAACUAAGAAAUGCAUUUAAAAUUAAACAAAUGCGCGCAAAAGUGCUGCAAUAAGUAAGACAUUAUG ..((.(((((((((...))))))))).)).......((((((..........))))))((((....))))..................... ( -21.00) >consensus UGGGUUGACAGAUGGCUCAUCUGUCAUCUAAUAAAUGCAUUUAAAAUUAAACAAAUGCGCGCAAAAGUGCUGCAAUAAGUUAGACAUUAUG ..((.(((((((((...))))))))).))......((((((((....)))).....((((......))))))))................. (-18.27 = -18.60 + 0.33)

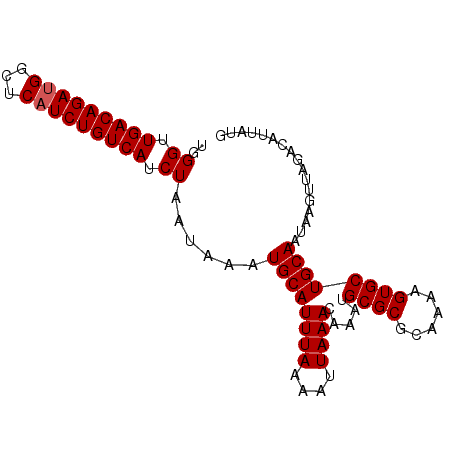
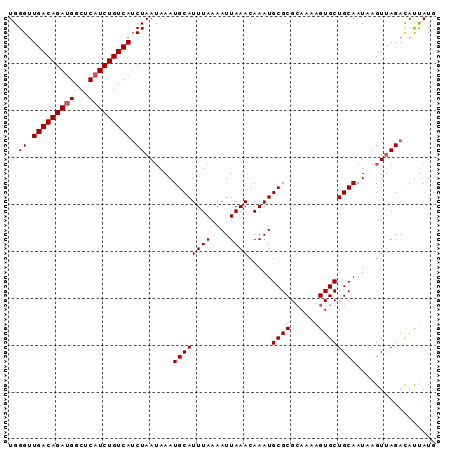
| Location | 1,254,278 – 1,254,369 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 91.58 |
| Mean single sequence MFE | -23.83 |
| Consensus MFE | -20.57 |
| Energy contribution | -21.13 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.42 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1254278 91 - 22407834 CAUAAUGUCUAACUGAUUGCAGCACUUAUGCGCGCAUUUGUUUAAUUUUAAAUGCAUUUAUUAGAUGACAGAUGAGCCGUCUGUCAACCCA ......((((((..(((.((.(((....)))))(((((((........))))))))))..))))))((((((((...))))))))...... ( -25.10) >DroSec_CAF1 14003 91 - 1 CAUGGUAUCUAACUUAUUGCAGCACUUCUUCGCGCAUUUGUUUAAUUUUAAAUGCAUUUAUUAGAUGACAGAUGAGCCAUCUGUCAACCCA ...(((((((((.........((........))(((((((........))))))).....))))))((((((((...)))))))).))).. ( -24.60) >DroSim_CAF1 14505 91 - 1 CAUAAUGUCUUACUUAUUGCAGCACUUUUGCGCGCAUUUGUUUAAUUUUAAAUGCAUUUCUUAGUUGACAGAUGAGCCAUCUGUCAUCCCA ..................((.(((....)))))(((((((........)))))))..........(((((((((...)))))))))..... ( -21.80) >consensus CAUAAUGUCUAACUUAUUGCAGCACUUAUGCGCGCAUUUGUUUAAUUUUAAAUGCAUUUAUUAGAUGACAGAUGAGCCAUCUGUCAACCCA ......((((((......((.(((....)))))(((((((........))))))).....))))))((((((((...))))))))...... (-20.57 = -21.13 + 0.56)

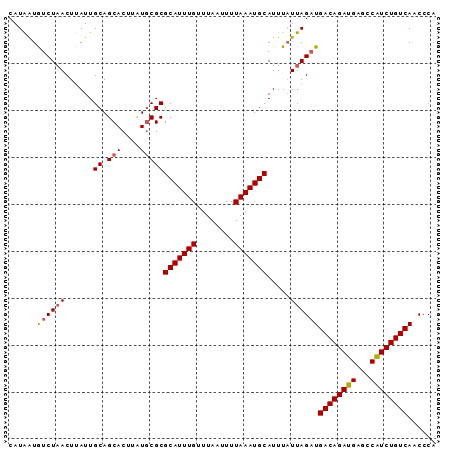
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:58 2006