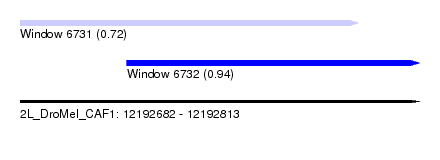
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,192,682 – 12,192,813 |
| Length | 131 |
| Max. P | 0.938402 |

| Location | 12,192,682 – 12,192,793 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 81.89 |
| Mean single sequence MFE | -23.17 |
| Consensus MFE | -12.42 |
| Energy contribution | -13.42 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.720256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
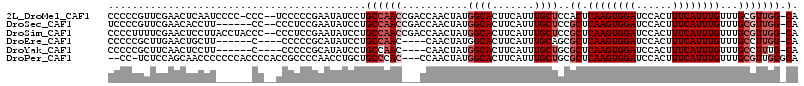
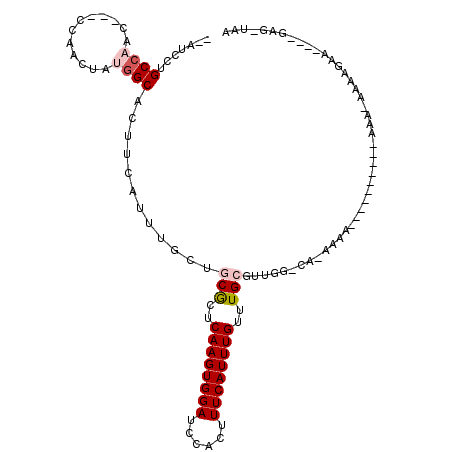
>2L_DroMel_CAF1 12192682 111 + 22407834 CCCCCGUUCGAACUCAAUCCCC-CCC--UCCCCCGAAUAUCCUGCCAACCGACCAACUAUGGCACUUCAUUUGCUCCACUCAAGUGGAUCCACUUUCAUUUGUUUGCGUUGG-CA .....(((((............-...--.....))))).....(((((((((.(((..((((((.......)))(((((....)))))........)))))).))).)))))-). ( -23.05) >DroSec_CAF1 21528 106 + 1 UCCCCGUUCGAACACCUU------CC--CCCUCCGAAUAUCCUGCCAACCGACCAACUAUGGCACUUCAUUUGCUCCGCUCAAGUGGAUCCACUUUCAUUUGUUUGCGUUGG-CA .....(((((........------..--.....))))).....((((((.(.(((....))))..............((.((((((((......))))))))...)))))))-). ( -23.66) >DroSim_CAF1 19459 112 + 1 CCCCUUUUCGAACUCCUUACCUACCC--CCCUCCGAAUAUCCUGCCAACCGACCAACUAUGGCACUUCAUUUGCUCCGCUCAAGUGGAUCCACUUUCAUUUGUUUGCGUUGG-CA ......((((................--.....))))......((((((.(.(((....))))..............((.((((((((......))))))))...)))))))-). ( -21.70) >DroEre_CAF1 19620 100 + 1 CCCCCGCUUGAACUGCUU------C----CCCCCGCAUAUCCUGCCAAC----CAACUAUGGCACUUCAUUUGCAGCGCUCAAGUGGAUCCACUUUCAUUUGUUUGCCUUGG-CA ...((((((((..((((.------.----.....(((.....(((((..----......))))).......))))))).)))))))).................(((....)-)) ( -23.41) >DroYak_CAF1 20215 100 + 1 CCCCCGCUUCAACUCCUU------C----CCCCCGCAUAUCCUGCCAAC----CAACUAUGGCACUUCAUUUGCUGCGCUCAAGUGGAUCCACUUUCAUUUGUUUGCCUUUG-CA .....((...........------.----.....(((.....(((((..----......))))).......))).(((..((((((((......))))))))..)))....)-). ( -20.60) >DroPer_CAF1 39495 109 + 1 --CC-UCUCCAGCAACCCCCCCACCCCACCGCCCCAACCUGCUGCCCAC---CCAACUAUGGCACUUCAUUUGCUGCGCUCAAGUGGAUCCACUUUCAUUUGUUUGCGUUGGGCA --..-....(((((.........................)))))....(---(((((...((((.......))))(((..((((((((......))))))))..))))))))).. ( -26.61) >consensus CCCCCGCUCGAACUCCUU______CC__CCCCCCGAAUAUCCUGCCAAC___CCAACUAUGGCACUUCAUUUGCUCCGCUCAAGUGGAUCCACUUUCAUUUGUUUGCGUUGG_CA ...........................................((((((...........((((.......))))..((.((((((((......))))))))...))))))).). (-12.42 = -13.42 + 1.00)
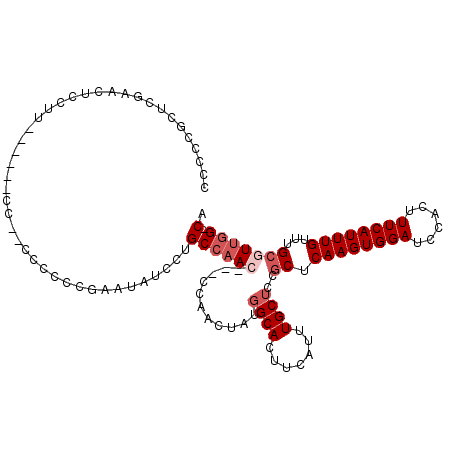
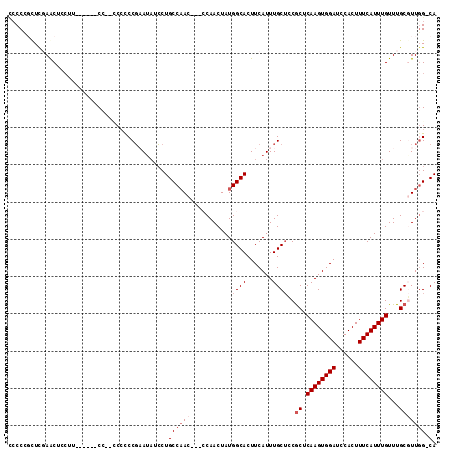
| Location | 12,192,717 – 12,192,813 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 77.94 |
| Mean single sequence MFE | -24.35 |
| Consensus MFE | -11.44 |
| Energy contribution | -12.13 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12192717 96 + 22407834 --AUCCUGCCAACCGACCAACUAUGGCACUUCAUUUGCUCCACUCAAGUGGAUCCACUUUCAUUUGUUUGCGUUGG-CA-AAAA----------AAA-AAAAGAA----GAG-UAA --....((((((((((.(((..((((((.......)))(((((....)))))........)))))).))).)))))-))-....----------...-.......----...-... ( -22.00) >DroGri_CAF1 24614 105 + 1 --CUCCAGC--AC-CAGCUCCUCUGGCACUUCAUUUGCAGCGCUCAAGUGGAUCCACUUUCAUUUGUUUGCGCACUGCAACAAAACGACACAA-AAAAGAAAGAA----AAA-AAA --....((.--.(-(((.....))))..))....(((((((((.((((((((......))))))))...))))..))))).............-...........----...-... ( -22.30) >DroSim_CAF1 19495 96 + 1 --AUCCUGCCAACCGACCAACUAUGGCACUUCAUUUGCUCCGCUCAAGUGGAUCCACUUUCAUUUGUUUGCGUUGG-CA-AAAA----------AAA-AAGAGUA----GAG-UAA --....(((((............)))))......((((((..((((((((....)))))...(((.(((((....)-))-)).)----------)).-..)))..----)))-))) ( -25.40) >DroYak_CAF1 20243 94 + 1 --AUCCUGCCAAC----CAACUAUGGCACUUCAUUUGCUGCGCUCAAGUGGAUCCACUUUCAUUUGUUUGCCUUUG-CA-AAAA----------AA---AGAGUAAAAUAAG-UAA --.....((((..----......))))((((..(((((((((..((((((((......))))))))..)))((((.-..-....----------))---))))))))..)))-).. ( -22.40) >DroAna_CAF1 20172 100 + 1 UCCUCCUGCCAAC----CAACUAUGGCACUUCAUUUGCUGCGCUCAAGUGGAUCCACUUUCAUUUGUUUGCUCUGGGCA-AAAA------CAAAAAA-AAAUGUA----GAGGAAA (((((.(((((..----......))))).....(((((((((..((((((((......))))))))..)))....))))-))..------.......-.......----))))).. ( -28.10) >DroPer_CAF1 39530 89 + 1 --CUGCUGCCCAC---CCAACUAUGGCACUUCAUUUGCUGCGCUCAAGUGGAUCCACUUUCAUUUGUUUGCGUUGGGCA-ACAA----------AAA-AAAAGA----------AA --.((.((((((.---........((((.......))))((((.((((((((......))))))))...))))))))))-.)).----------...-......----------.. ( -25.90) >consensus __AUCCUGCCAAC___CCAACUAUGGCACUUCAUUUGCUGCGCUCAAGUGGAUCCACUUUCAUUUGUUUGCGUUGG_CA_AAAA__________AAA_AAAAGAA____GAG_UAA .......((((............))))............(((..((((((((......))))))))..)))............................................. (-11.44 = -12.13 + 0.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:47 2006