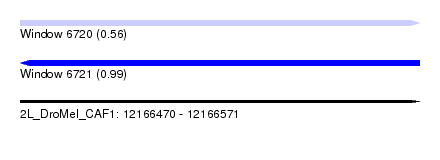
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,166,470 – 12,166,571 |
| Length | 101 |
| Max. P | 0.994177 |

| Location | 12,166,470 – 12,166,571 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 74.83 |
| Mean single sequence MFE | -28.95 |
| Consensus MFE | -12.91 |
| Energy contribution | -13.13 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
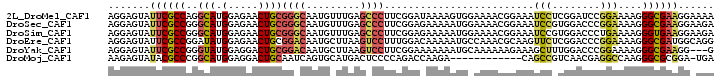
>2L_DroMel_CAF1 12166470 101 + 22407834 AGGAGUAUUCGCCAGGCAUGGAGAACUGCGGGCAAUGUUUGAGCCCUUCGGAUAAAAGUGGAAAACGGAAAUCCUCGGAUCCGGAAAAGGGCGAAGGAAAA .......((((((..(((.(.....))))((((.........))))(((((((....(.(((.........))).)..)))))))....))))))...... ( -29.40) >DroSec_CAF1 51501 101 + 1 AGGAGUAUUCGCCGGGCAUGGAGAACUGCGGGCAAUGUUUGAGCCCUUCGGAGAAAAAUGGAAAACGGAAAUCCGUGGACCCGGAAAAGGGCGAAGGAAGA ....((.((..(((((((((((...(((.((((.........))))((((........))))...)))...))))))..)))))..))..))......... ( -27.90) >DroSim_CAF1 49116 101 + 1 AGGAGUAUUCGCCGGGCAUGGAGAACUGCGGGCAAUGUUUGAGCCCUUCGGAGAAAAAUGGAAAACGGAAAUCCGUGGACCCUGAAAAGGGUGAAGGAAGA .......(((.(((((((.(.....))))((((.........)))).)))).))).........((((....))))..(((((....)))))......... ( -29.70) >DroEre_CAF1 47619 101 + 1 AGGAGUAUUCGCCGGAUAUGGAGAACUGCGGACAAUGCUUAAGUCCUUUGGACAAAAAUGCCAAACGCAAGUUCUCGGACCCGGAAAAGGGCGAUGGCAGG ..........((((......(((((((((((((.........))))(((((.((....))))))).)).))))))).(.(((......))))..))))... ( -31.90) >DroYak_CAF1 42579 98 + 1 AGGAGUAUUCGCCGGGUAUGGAGGACUGCGGACAAUGCUUAAGUCCUUCGGAAAAAAAUGCAAAAAAGAAAGCUUUGGACCCGGAAAAGGGCGAAGG---G ....((.((..((((((.((((((((((((.....)))...)))))))))..............((((....))))..))))))..))..)).....---. ( -27.00) >DroMoj_CAF1 56354 88 + 1 AAGAGUAUACGCCCGGCAUGGAGGACUGCAAUCAGUGCAUGACUCCCCAGACCAAGA------------CAGCCGUCAACGAGGCCAAGGGCGCGGA-UGA .........(((((((..(((.(((.((((.....))))....))))))..))....------------..(((........)))...)))))....-... ( -27.80) >consensus AGGAGUAUUCGCCGGGCAUGGAGAACUGCGGGCAAUGCUUGAGCCCUUCGGACAAAAAUGGAAAACGGAAAUCCGUGGACCCGGAAAAGGGCGAAGGAAGA .......((((((..(((.(.....))))((((.........)))).........................(((........)))....))))))...... (-12.91 = -13.13 + 0.23)
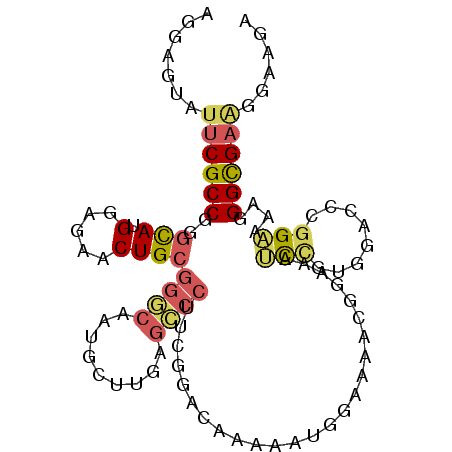
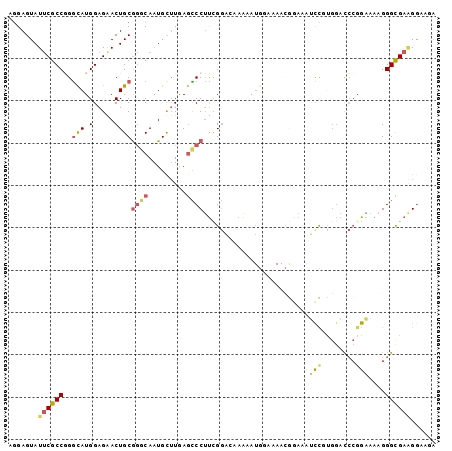
| Location | 12,166,470 – 12,166,571 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 74.83 |
| Mean single sequence MFE | -27.58 |
| Consensus MFE | -12.83 |
| Energy contribution | -13.03 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.47 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12166470 101 - 22407834 UUUUCCUUCGCCCUUUUCCGGAUCCGAGGAUUUCCGUUUUCCACUUUUAUCCGAAGGGCUCAAACAUUGCCCGCAGUUCUCCAUGCCUGGCGAAUACUCCU .........((((((...(((((..(((...............)))..))))))))))).......(((((.(((........)))..)))))........ ( -25.16) >DroSec_CAF1 51501 101 - 1 UCUUCCUUCGCCCUUUUCCGGGUCCACGGAUUUCCGUUUUCCAUUUUUCUCCGAAGGGCUCAAACAUUGCCCGCAGUUCUCCAUGCCCGGCGAAUACUCCU ......((((((.......(((...((((....))))..))).............((((......((((....)))).......))))))))))....... ( -26.82) >DroSim_CAF1 49116 101 - 1 UCUUCCUUCACCCUUUUCAGGGUCCACGGAUUUCCGUUUUCCAUUUUUCUCCGAAGGGCUCAAACAUUGCCCGCAGUUCUCCAUGCCCGGCGAAUACUCCU ...(((((((((((....)))))..((((....))))...............))))))........(((((.(((........)))..)))))........ ( -25.90) >DroEre_CAF1 47619 101 - 1 CCUGCCAUCGCCCUUUUCCGGGUCCGAGAACUUGCGUUUGGCAUUUUUGUCCAAAGGACUUAAGCAUUGUCCGCAGUUCUCCAUAUCCGGCGAAUACUCCU ..((((...((((......))))..(((((((.((.(((((((....)).)))))((((.........))))))))))))).......))))......... ( -33.50) >DroYak_CAF1 42579 98 - 1 C---CCUUCGCCCUUUUCCGGGUCCAAAGCUUUCUUUUUUGCAUUUUUUUCCGAAGGACUUAAGCAUUGUCCGCAGUCCUCCAUACCCGGCGAAUACUCCU .---..((((((......((((...(((((..........)).)))...)))).((((((...((.......))))))))........))))))....... ( -21.30) >DroMoj_CAF1 56354 88 - 1 UCA-UCCGCGCCCUUGGCCUCGUUGACGGCUG------------UCUUGGUCUGGGGAGUCAUGCACUGAUUGCAGUCCUCCAUGCCGGGCGUAUACUCUU ...-...((((((..((((........)))).------------....(((.(((((.(.(.((((.....))))).)))))).)))))))))........ ( -32.80) >consensus UCUUCCUUCGCCCUUUUCCGGGUCCAAGGAUUUCCGUUUUCCAUUUUUCUCCGAAGGACUCAAACAUUGCCCGCAGUUCUCCAUGCCCGGCGAAUACUCCU .......(((((.......((((((..(((...................)))...))))))....((((....))))...........)))))........ (-12.83 = -13.03 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:37 2006