| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,150,714 – 12,150,817 |
| Length | 103 |
| Max. P | 0.779517 |

| Location | 12,150,714 – 12,150,817 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.58 |
| Mean single sequence MFE | -27.52 |
| Consensus MFE | -16.59 |
| Energy contribution | -16.20 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
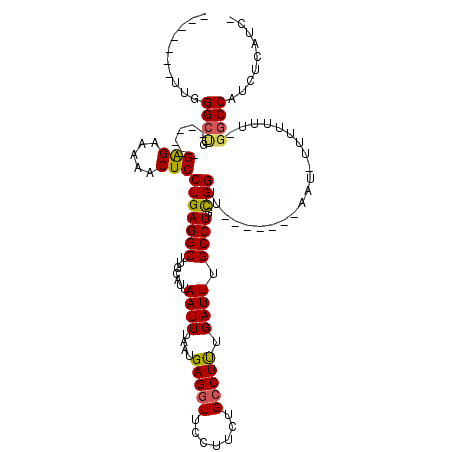
>2L_DroMel_CAF1 12150714 103 - 22407834 UUUUUUUUUGGGCUG------GAGAAAAACUCCUGAGGCUUGCAUUAAUUUAAUGAGGUUCCUUCUGCCUUUGAUUUGCCUGUGGUU-------AAU-UU-UUUU-GGCCAUCACAUCG ..........(((..------((((((....((..((((...(((((...)))))((((.......)))).......))))..))..-------..)-))-))).-.)))......... ( -25.10) >DroPse_CAF1 80303 102 - 1 --------UGGGCCGUGAGGAGGGAAAAUCUCCUGAGGCCUGCAUUAAUUUAAUGAGUUUCCUUCUGCCUCUGAUUUGCCUGCGGUU-------UUCGUUUUUUU-GGCCAUUUCAUU- --------..(((((.((((((.(((((((((..(((((.......(((((...))))).......))))).))...(....)))))-------))).)))))))-))))........- ( -26.14) >DroSec_CAF1 35981 99 - 1 -----UUUUUGGCUG------GAGAAAAACUCCUGAGGCUUGCAUUAAUUUAAUGAGGUUCCUUCUGCCUUUGAUUUGCCUGUGGUU-------AAU-UUUUUUU-GGCCAUGCCAUCG -----....((((.(------(((.....))))..((((...(((((...)))))((((.......)))).......))))((((((-------((.-.....))-))))))))))... ( -26.90) >DroYak_CAF1 26434 102 - 1 --UUUUUUUGGGCUG------GGGAAAAACUCCUGAGGCUUGCAUUAAUUUAAUGAGGUUUCUGCUGCCUUUGAUUUGCCUGUGGUU-------AAU-UUUUUUC-GGCCAUCCCAUCA --........(((((------..(((((...((..((((...(((((...)))))((((.......)))).......))))..))..-------..)-))))..)-))))......... ( -27.20) >DroAna_CAF1 30587 95 - 1 -------UAGGGGAG------GAGG-AAACUCCUGAGGCUUGCAUUAAUUUAAUGAGGUUCCUUCUGCCUUUGAUUUGCCUGCGGCU-------CCU-UUUUUUCAGACCAUCUCAU-- -------.(((((((------(((.-...)))))...((..(((..((((....(((((.......))))).)))))))..))..))-------)))-...................-- ( -26.20) >DroPer_CAF1 80451 109 - 1 --------UGGGCCGUGAGGAGGGAAAAUCUCCUGAGGCCUGCAUUAAUUUAAUGAGGUUCCUUCUGCCUCUGAUUUGCCUGCGGUUUUCGGUUUUCGUUUUUUU-GGCCAUUUCAUU- --------..(((((.((((((.(((((((....((((((.(((..((((....(((((.......))))).))))....))))))))).))))))).)))))))-))))........- ( -33.60) >consensus _______UUGGGCUG______GAGAAAAACUCCUGAGGCUUGCAUUAAUUUAAUGAGGUUCCUUCUGCCUUUGAUUUGCCUGCGGUU_______AAU_UUUUUUU_GGCCAUCUCAUC_ ..........((((.......(((.....)))(((((((.......((((....(((((.......))))).)))).)))).))).....................))))......... (-16.59 = -16.20 + -0.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:30 2006