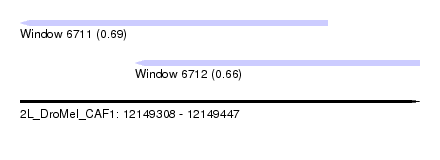
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,149,308 – 12,149,447 |
| Length | 139 |
| Max. P | 0.685802 |

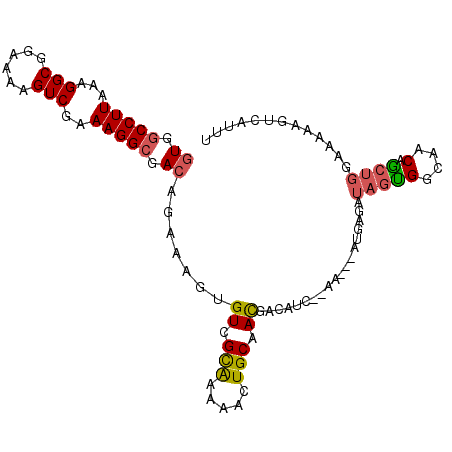
| Location | 12,149,308 – 12,149,415 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 88.87 |
| Mean single sequence MFE | -25.26 |
| Consensus MFE | -19.00 |
| Energy contribution | -18.48 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12149308 107 - 22407834 CAGAAAGUGUCGCAAAAACUGCAACGACAUC--AAAUGAGAUAGUGGCAACAGCUGGAAAAAGUCAUUUCGCCUCCAGCACUCAGAGCAACAUUAGAAACAGACGCACA ......((((((((.....))).........--...((((....((....))((((((....(......)...)))))).)))).................)))))... ( -25.20) >DroSec_CAF1 34669 107 - 1 CAGAAAGUGUCGCAAAAACUGCAACGACAUC--AAAUGAGAUAGUGGCAACAGCUGGAAAAAGUCAUUUUGCCUCCAGCACUCAGAGCAACAUUAGAAACAGACGCACA ......((((((((.....))).........--...((((....((....))((((((..(((....)))...)))))).)))).................)))))... ( -25.80) >DroSim_CAF1 31296 107 - 1 CAGAAAGUGUCGCAAAAACUGCAACGACAUC--AAAUGAGAUAGUGGCAACAGCUGGAAAAAGUCAUUUUGCCUCCAGCACUCAGAGCAACAUUAGAAACAGACGCACA ......((((((((.....))).........--...((((....((....))((((((..(((....)))...)))))).)))).................)))))... ( -25.80) >DroEre_CAF1 30449 107 - 1 CAAAAAGUGUCGUAGAAACUGCAACGACAUC--AAAUGAGAUAGCGGCCACAGCUGGGAAAAGUCAUUUUGCUCCCGGCUCUAAGAGCCACAUUAGAAGCAGAUGCACA ......(((.(((.....((((...(..(((--......)))..)((((..((((((((.(((....)))..))))))))....).))).........)))))))))). ( -27.30) >DroYak_CAF1 25024 109 - 1 CAGAAAGUGUCGUAGAAACUGCAACUGAAACUGCUAUGAGAUAGCGACAACAGCUGGGAAAAGUCAUUUCGCUUCGGGCACUAAGAGCCACAUUAGAAACAGAUGCACA (((....((((((((...))))..........(((((...)))))))))....)))......(((((((.(.(((.(((.......)))......))).)))))).)). ( -22.20) >consensus CAGAAAGUGUCGCAAAAACUGCAACGACAUC__AAAUGAGAUAGUGGCAACAGCUGGAAAAAGUCAUUUUGCCUCCAGCACUCAGAGCAACAUUAGAAACAGACGCACA ......((((((((.....)))..............((((.....(....).((((((...............)))))).)))).................)))))... (-19.00 = -18.48 + -0.52)

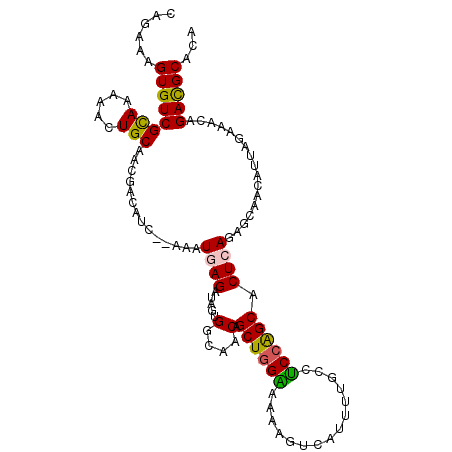
| Location | 12,149,348 – 12,149,447 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 80.20 |
| Mean single sequence MFE | -24.33 |
| Consensus MFE | -14.21 |
| Energy contribution | -14.18 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12149348 99 - 22407834 GUGGCCUUAAAGGCGGAAAAGUCGAAAGGCAACAGAAAGUGUCGCAAAAACUGCAACGACAUC--AA---AUGAGAUAGUGGCAACAGCUGGAAAAAGUCAUUU ...(((((...(((......)))..)))))..(((....(((((((.....)))....(((((--..---....))).))))))....)))............. ( -21.80) >DroSec_CAF1 34709 99 - 1 GUGUCCUUAAAGGCGGAAAAGUCGAAAGGCGACAGAAAGUGUCGCAAAAACUGCAACGACAUC--AA---AUGAGAUAGUGGCAACAGCUGGAAAAAGUCAUUU (((((.......((((......(....)((((((.....)))))).....))))...))))).--((---((((..(((((....)).))).......)))))) ( -22.90) >DroSim_CAF1 31336 99 - 1 GUGGCCUUAAAGGCGGAAAAGUCGAAAGGCGACAGAAAGUGUCGCAAAAACUGCAACGACAUC--AA---AUGAGAUAGUGGCAACAGCUGGAAAAAGUCAUUU ((.(((((...(((......)))..))))).)).....((((((((.....)))...))))).--((---((((..(((((....)).))).......)))))) ( -23.60) >DroEre_CAF1 30489 99 - 1 GUGGCCUUAAAGGCGGAAAAGUCGAAAGGCGACAAAAAGUGUCGUAGAAACUGCAACGACAUC--AA---AUGAGAUAGCGGCCACAGCUGGGAAAAGUCAUUU ((((((......((((....(((....)))((((.....)))).......))))...(..(((--..---....)))..))))))).(((......)))..... ( -28.80) >DroYak_CAF1 25064 101 - 1 GUGGCCUUAAAGGCGGAAAAGUCGAAAGGCGACAGAAAGUGUCGUAGAAACUGCAACUGAAACUGCU---AUGAGAUAGCGACAACAGCUGGGAAAAGUCAUUU (((((((((..(((((....(((....)))((((.....))))((((...))))........)))))---.)))).((((.......))))......))))).. ( -25.10) >DroAna_CAF1 29246 85 - 1 UUGGCCUUUAAGGCG-ACAGGUCGAAAGGCAACAGGAGCUGUCGCGUCACCUGCAAUGACACG--AAGGAAGGAAAUAUUGCAAACGA---------------- ....(((((...(((-((((.((....(....)..)).)))))))((((.......))))...--...)))))...............---------------- ( -23.80) >consensus GUGGCCUUAAAGGCGGAAAAGUCGAAAGGCGACAGAAAGUGUCGCAAAAACUGCAACGACAUC__AA___AUGAGAUAGUGGCAACAGCUGGAAAAAGUCAUUU ((.(((((...(((......)))..))))).)).......((.(((.....))).))...................(((((....).))))............. (-14.21 = -14.18 + -0.02)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:28 2006