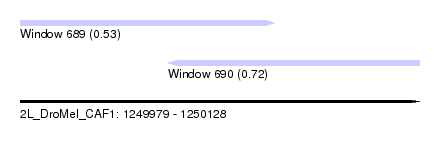
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,249,979 – 1,250,128 |
| Length | 149 |
| Max. P | 0.721324 |

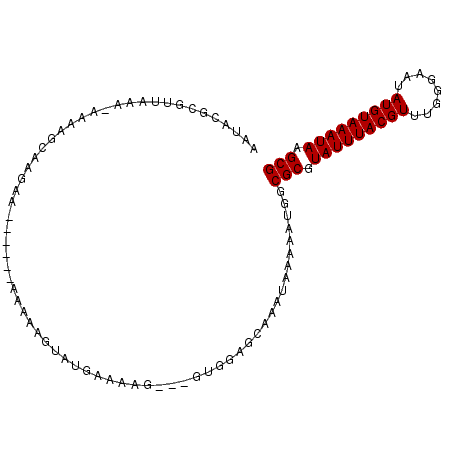
| Location | 1,249,979 – 1,250,074 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 79.43 |
| Mean single sequence MFE | -14.37 |
| Consensus MFE | -9.97 |
| Energy contribution | -10.17 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1249979 95 + 22407834 GCCAAAAGA---CUUU--GUCCGGAUGCCACGU--CAAGCUAAAGAAAACAAAUACAACGGACGCUUAUUUACAUAUUCCCAAACGUAAAUACGCGCCAUUU .........---((((--(.(..((((...)))--)..).)))))..............((.(((.(((((((............))))))).))))).... ( -16.80) >DroVir_CAF1 28114 83 + 1 ---AAA----------------AGACGCAACGGCACAAGCUAAAGGAAAUAAAUACAACGGACUCUUAUUUACAUAUUCCCAAACGUAAAUACGCGCCAUUU ---...----------------.(.(((...(((....))).........................(((((((............))))))).))).).... ( -9.30) >DroPse_CAF1 14752 100 + 1 CCCAAGACUCCCCUUUUCGCUGGGACGCCACGU--AAAGCUAAAGAAAAUAAAUACAACGGACGCUUAUUUACAUAUUCCCAAACGUAAAUACGCGCCAUUU ............((((..(((...(((...)))--..))).))))..............((.(((.(((((((............))))))).))))).... ( -17.80) >DroWil_CAF1 13867 81 + 1 -------------------AGACGACUCCACAU--CAACCUAAAGAAAAUAAAUACAACGGACGCUUAUUUGCAUAUUCCCAAACGUAAAUACGCACCAUUU -------------------..............--........................((..((.(((((((............))))))).)).)).... ( -7.90) >DroYak_CAF1 10381 95 + 1 A---GCACCCCCCUUU--GUCCGGAUGCCACGU--CAAGCUAAAGAAAACAAAUACAACGGACGCUUAUUUACAUAUUCCCAAACGUAAAUACGCGCCAUUU .---........((((--(.(..((((...)))--)..).)))))..............((.(((.(((((((............))))))).))))).... ( -16.60) >DroPer_CAF1 14607 100 + 1 CCCAAGACUCCCCUUUUCGCUGGGACGCCACGU--AAAGCUAAAGAAAAUAAAUACAACGGACGCUUAUUUACAUAUUCCCAAACGUAAAUACGCGCCAUUU ............((((..(((...(((...)))--..))).))))..............((.(((.(((((((............))))))).))))).... ( -17.80) >consensus ___AAAAC____CUUU__GCCCGGACGCCACGU__CAAGCUAAAGAAAAUAAAUACAACGGACGCUUAUUUACAUAUUCCCAAACGUAAAUACGCGCCAUUU ...........................................................((.(((.(((((((............))))))).))))).... ( -9.97 = -10.17 + 0.20)

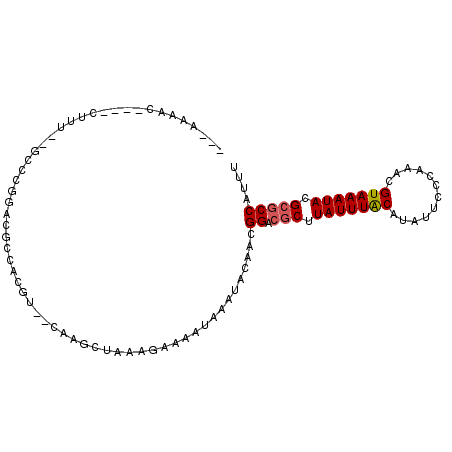
| Location | 1,250,034 – 1,250,128 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 83.13 |
| Mean single sequence MFE | -13.74 |
| Consensus MFE | -10.60 |
| Energy contribution | -10.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1250034 94 - 22407834 AAUACGCGUUAAA-AAAGGCAAAAA----AAAAAAAGUAUGAAAAG---GUGGAGCAAAUAAAAAUGGCGCGUAUUUACGUUUGGGAAUAUGUAAAUAAGCG ..(((((((((..-....((.....----.................---.....)).........)))))))))(((((((........)))))))...... ( -13.09) >DroSec_CAF1 9836 92 - 1 AAUACGCGUUAAA-AAAAGCAAGAA------AAAAUGUAUGAAAAG---GUGGAGCAAAUAAAAAUGGCGCGUAUUUACGUUUGGGAAUAUGUAAAUAAGCG ..(((((((((..-....(((....------....)))........---(.....).........)))))))))(((((((........)))))))...... ( -13.90) >DroSim_CAF1 10316 93 - 1 AAUACGCGUUAAAAAAAAGCAAGAA------AAAAUGUAUGAAAAG---GUGGAGCAAAUAAAAAUGGCGCGUAUUUACGUUUGGGAAUAUGUAAAUAAGCG ..(((((((((.......(((....------....)))........---(.....).........)))))))))(((((((........)))))))...... ( -13.90) >DroEre_CAF1 22957 85 - 1 ------CGUUAAA-AACUGGAAGGA-------AAAAGUACAAAAAG---GUGGAGCAAAUAAAAAUGGCGCGUAUUUACGUUUGGGAAUAUGUAAAUAAGCG ------.(((...-.(((.....(.-------.......).....)---))..)))............(((.(((((((((........))))))))).))) ( -12.30) >DroAna_CAF1 33184 100 - 1 AAUAUACAUUGAA-AC-AAAAAGAAACGAAGGACAAAAAUAAAAAGCUCGUGCUGCAAAUAAAAAUGGCGCGUAUUUACGUUUGGGAAUAUGUAAAUAAGCG .............-..-.........(....).............(((((((((............))))))(((((((((........)))))))))))). ( -15.50) >consensus AAUACGCGUUAAA_AAAAGCAAGAA______AAAAAGUAUGAAAAG___GUGGAGCAAAUAAAAAUGGCGCGUAUUUACGUUUGGGAAUAUGUAAAUAAGCG ....................................................................(((.(((((((((........))))))))).))) (-10.60 = -10.60 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:54 2006