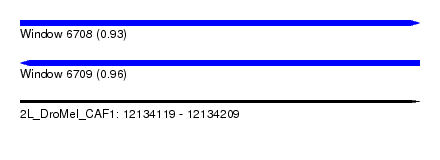
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,134,119 – 12,134,209 |
| Length | 90 |
| Max. P | 0.962685 |

| Location | 12,134,119 – 12,134,209 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 81.62 |
| Mean single sequence MFE | -23.73 |
| Consensus MFE | -18.82 |
| Energy contribution | -18.60 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12134119 90 + 22407834 AUGUGGUGAGAAGCACUCGCUCUCUUGGCAGUAGCGAUUAAGCUUUCAGAGAGAGCUUUUU--UUUACUCAUUUCAAAGUUUAUGCUUUAAC ..(..((((((((.....((((((((......(((......)))....)))))))).....--))).)))))..)(((((....)))))... ( -23.60) >DroSec_CAF1 14458 90 + 1 AUGUGGUGAGAAGCGCUCGCUCUCUUGGCAGUAGCGAUUAAGCUUUCAGAGAGAGCUUUUUGCUUUACUCAUUUCAAAGUUUAUGCUUAA-- .((..((((((((((...((((((((......(((......)))....))))))))....)))))..)))))..))((((....))))..-- ( -27.90) >DroSim_CAF1 15495 70 + 1 AAGUGGUGAGAAGCGCUCGCUCUCUUGGCAGUAGCGAUUAAGCUUUCAGAGAGAGCUUUU--------------------UUAUGCUUUA-- ..((.(.((((.((....)))))).).))((((..((..((((((((...))))))))..--------------------)).))))...-- ( -19.70) >consensus AUGUGGUGAGAAGCGCUCGCUCUCUUGGCAGUAGCGAUUAAGCUUUCAGAGAGAGCUUUUU__UUUACUCAUUUCAAAGUUUAUGCUUUA__ ..(((.(....).)))..((((((((......(((......)))....)))))))).................................... (-18.82 = -18.60 + -0.22)



| Location | 12,134,119 – 12,134,209 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 81.62 |
| Mean single sequence MFE | -18.67 |
| Consensus MFE | -15.97 |
| Energy contribution | -16.30 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12134119 90 - 22407834 GUUAAAGCAUAAACUUUGAAAUGAGUAAA--AAAAAGCUCUCUCUGAAAGCUUAAUCGCUACUGCCAAGAGAGCGAGUGCUUCUCACCACAU ....((((((..((((......))))...--.....(((((((..(..(((......)))....)..)))))))..)))))).......... ( -20.90) >DroSec_CAF1 14458 90 - 1 --UUAAGCAUAAACUUUGAAAUGAGUAAAGCAAAAAGCUCUCUCUGAAAGCUUAAUCGCUACUGCCAAGAGAGCGAGCGCUUCUCACCACAU --...................((((..((((.....(((((((..(..(((......)))....)..)))))))....))))))))...... ( -20.00) >DroSim_CAF1 15495 70 - 1 --UAAAGCAUAA--------------------AAAAGCUCUCUCUGAAAGCUUAAUCGCUACUGCCAAGAGAGCGAGCGCUUCUCACCACUU --..........--------------------....(((((((..(..(((......)))....)..)))))))(((.....)))....... ( -15.10) >consensus __UAAAGCAUAAACUUUGAAAUGAGUAAA__AAAAAGCUCUCUCUGAAAGCUUAAUCGCUACUGCCAAGAGAGCGAGCGCUUCUCACCACAU ....((((....((((......))))..........(((((((..(..(((......)))....)..)))))))....)))).......... (-15.97 = -16.30 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:25 2006