
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,119,744 – 12,119,910 |
| Length | 166 |
| Max. P | 0.649431 |

| Location | 12,119,744 – 12,119,855 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.99 |
| Mean single sequence MFE | -31.08 |
| Consensus MFE | -21.96 |
| Energy contribution | -21.68 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12119744 111 - 22407834 C----CCAUCCAAAGUUAUUAGAGCCGCGAGGGCUCAAUAAAUGGGCGUCGUUUAUGCACCUCAUCAUCAGCGAUUGCACAGAUCGCAGC-----UCUGUGUUCUUGCCUCUUUUUUUUU .----.......(((....((((((...(((((((((.....)))))...((....)).)))).......((((((.....)))))).))-----))))....))).............. ( -29.20) >DroPse_CAF1 30989 107 - 1 -----GCGUACAAAGUUAUUAGAGCCGCGAGGGCUCAAUAAAUGGGCGUCGUUUAUGCACCUCAUCAUCAGCGAUUGCACAGAUCGCAGAGUUCUUCUCGCUUGUUGCCUGU-------- -----(((.((((.((....((((((..(((((((((.....)))))...((....)).)))).......((((((.....)))))).).)))))....)))))))))....-------- ( -30.40) >DroSec_CAF1 208 109 - 1 C----CCAACCAAAGUUAUUAGAGCCGCGAGGGCCCAAUAAAUGGGCGUCGUUUAUGCACCUCAUCAUCAGCGAUUGCACAGAUCGCAGC-----UCUGUAUUCUUGCCUUUU--UUUUA .----.......(((....((((((...(((((((((.....)))))...((....)).)))).......((((((.....)))))).))-----))))....))).......--..... ( -32.20) >DroEre_CAF1 205 104 - 1 C----CCAUCCAAAGUUAUUAGAGCCGCGAGGGCUCAAUAAAUGGGCGUCGUUUAUGCACCUCGUCAUCGGCGAUUGCACAGAUCGCAGC-----UCCGUGUUUUUGCUUUUU------- .----.....(((((.(((..((((.(((((((((((.....)))))...((....)).)))))).....((((((.....)))))).))-----)).))).)))))......------- ( -29.40) >DroAna_CAF1 182 102 - 1 CCUCCCUCCCAGCAGCCAUUGGGCCCGCGAGGGCUCAAUAAAUGGGCGUCGUUUAUGCACCUCAUCAUCAGCGAUUGCACAGAUCGCGGU------------UUUUGUUUUU------UA .........(((.(((((((((((((....)))))))))....((((((.....)))).)).........((((((.....)))))))))------------).))).....------.. ( -34.90) >DroPer_CAF1 30862 108 - 1 -----GCGUACAAAGUUAUUAGAGCCGCGAGGGCUCAAUAAAUGGGCGUCGUUUAUGCACCUCAUCAUCAGCGAUUGCACAGAUCGCAGAGUUCUUCUCGCUUGUUGCCUGUU------- -----(((.((((.((....((((((..(((((((((.....)))))...((....)).)))).......((((((.....)))))).).)))))....))))))))).....------- ( -30.40) >consensus C____CCAUCCAAAGUUAUUAGAGCCGCGAGGGCUCAAUAAAUGGGCGUCGUUUAUGCACCUCAUCAUCAGCGAUUGCACAGAUCGCAGC_____UCUGUCUUCUUGCCUUUU_______ ..........................(((((((((((.....)))))...((....)).)))).......((((((.....))))))...................))............ (-21.96 = -21.68 + -0.28)


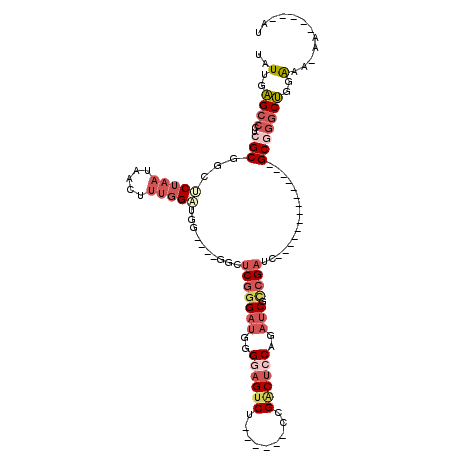
| Location | 12,119,819 – 12,119,910 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.62 |
| Mean single sequence MFE | -39.79 |
| Consensus MFE | -14.38 |
| Energy contribution | -17.06 |
| Covariance contribution | 2.68 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.36 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12119819 91 + 22407834 UAUUGAGCCCUCGCGGCUCUAAUAACUUUGGAUGG----GGAUCGGGUUGGGGAGUCU------CCGACUGCAGAUCGACGAUC--------------GCGGGCUGGAAAAAA-----AU ..((.(((((..((((.(((((.....)))))((.----.((((.((((((((...))------))))))...))))..)).))--------------))))))).)).....-----.. ( -31.40) >DroSec_CAF1 281 90 + 1 UAUUGGGCCCUCGCGGCUCUAAUAACUUUGGUUGG----GGCUCGGGAUGGGGAGUCU------CCGACUCCAGAUCGCCGAUC--------------GCGCGCCUGAAA-AA-----AU ..(..(((...((((((((((((.......)))))----)))(((((((..((((((.------..))))))..))).)))).)--------------))).)))..)..-..-----.. ( -39.00) >DroSim_CAF1 286 90 + 1 UAUUGAGCCCUCGCGGCUCUAAUAACUUUGGUUGG----GGCUCGGGAUGGGGAGUCU------CCGACUCCAGAUCGCCGAUC--------------GCGGGCUGGAAA-AA-----AU ..((.(((((..(((((((((((.......)))))----)))(((((((..((((((.------..))))))..))).)))).)--------------))))))).))..-..-----.. ( -40.10) >DroEre_CAF1 273 110 + 1 UAUUGAGCCCUCGCGGCUCUAAUAACUUUGGAUGG----GGUGCGGGAUGGGGAGUCC------CCGGCUCCAGAUCGUCGAUCGCCGAUCGUCGAUCGCCGGCUGGAAAUGGAGGUGGA ....(((((((((((.(((..............))----).))))))..((((...))------)))))))..(((((.(((((...))))).))))).(((.((........)).))). ( -44.04) >DroAna_CAF1 244 105 + 1 UAUUGAGCCCUCGCGGGCCCAAUGGCUGCUGGGAGGGAGGGGUCUGGAGCGGAUGACUCUGGAGCUGCCUCCAGAUCGUUGAUC--------------GCUGACUUGGCCCAAAGG-GGC ....((.(((((.(...((((........)))).).))))).))...(((((((((.(((((((....))))))))))))...)--------------)))......((((....)-))) ( -44.40) >consensus UAUUGAGCCCUCGCGGCUCUAAUAACUUUGGAUGG____GGCUCGGGAUGGGGAGUCU______CCGACUCCAGAUCGCCGAUC______________GCGGGCUGGAAA_AA_____AU ..(..((((..(((...(((((.....)))))..........(((((((..((((((.........))))))..))).))))................)))))))..)............ (-14.38 = -17.06 + 2.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:19 2006