
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,117,853 – 12,118,124 |
| Length | 271 |
| Max. P | 0.979160 |

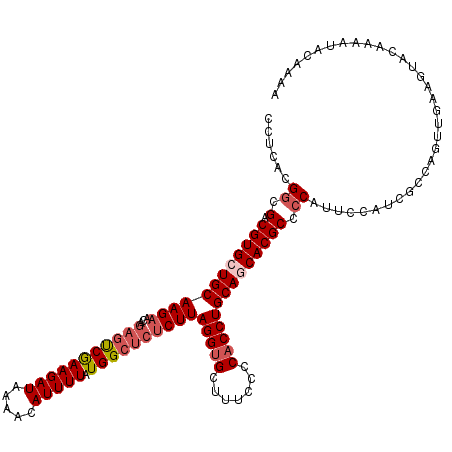
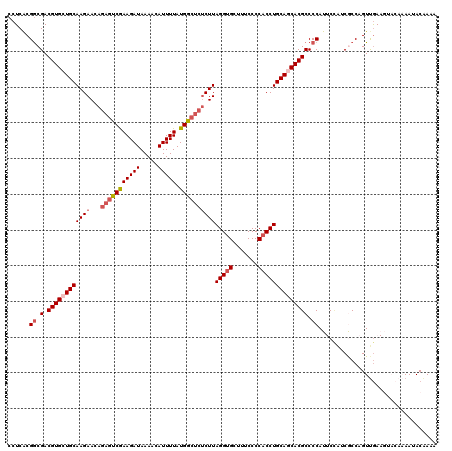
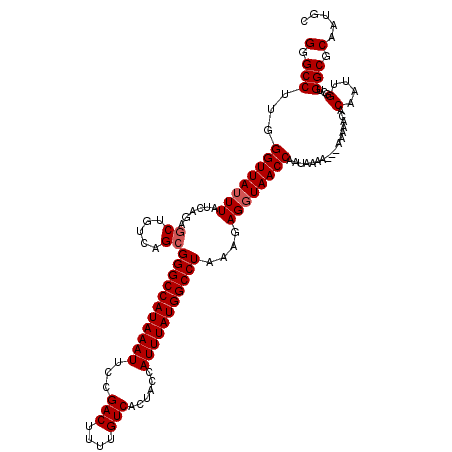
| Location | 12,117,853 – 12,117,973 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.16 |
| Mean single sequence MFE | -29.58 |
| Consensus MFE | -25.26 |
| Energy contribution | -26.54 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12117853 120 + 22407834 CCUCACGGCGACGUGCUGCAAGAACAGAGUCGAAGAUAAAACAUUUUAUGGCUCUCUUAGGUGCUUUCCCCACCUGCAGCACGCCCCAUUCCAACGCCAGUUGAAGUACAAAAUACAAAA ......((.(.((((((((((((...(((((((((((.....))))).))))))))))(((((.......))))))))))))).)))....((((....))))................. ( -35.50) >DroSec_CAF1 19993 120 + 1 CCUCACGGCGACGUGCUGCAAGAACAGAGCCGAAGAUAAAACAUUUUAUGGCUCACUUAGGUGCUUUCCCCACCUGCAGCACGCCCCAUUCCGUCGCCAGUUGAAGUACAAAAUACAAAA ......(((((((((((((.......(((((((((((.....))))).))))))....(((((.......))))))))))))(........)))))))...................... ( -40.10) >DroSim_CAF1 19874 120 + 1 CCUCGCGGCGACGUGCUGCAAGAACAGACUCGAAGAUAAAACAUUUUAUGGCUCUCUUAGGUGCUUUCCCCACCUGCAGCACGCCCCAUUCCAUCGCCAGUUGAAGUACAAAAUACAAAA ......(((((((((((((((((..((.((..(((((.....)))))..)))).))))(((((.......)))))))))))))..........)))))...................... ( -30.00) >DroEre_CAF1 20267 109 + 1 CCUCACGACGACGUGCUGCAAGAAAAAAGUCAAAGAUAAAAAAUUUUAUGGCUCUCUUAGGUGAAUUCCCCACCUGCAACACGCCCCAU-C---CACCAGUCAA-------AAGACAAAA ...........((((.(((((((....((((((((((.....))))).))))).))))(((((.......)))))))).))))......-.---.....(((..-------..))).... ( -22.50) >DroYak_CAF1 20190 112 + 1 CCUCACGGCGACGUGCUGCAAGAACAGCGUCGAAGAUAAAACAUUUUAUGGCUCUCUUAGGAGAUUUCCCCACCUGCAACACGCCCCAU-CUACCACCAGUUGA-------AAUACGAAA ......((.(.((((.((((.(......(((((((((.....))))).))))((((....))))........).)))).)))).)))..-..............-------......... ( -19.80) >consensus CCUCACGGCGACGUGCUGCAAGAACAGAGUCGAAGAUAAAACAUUUUAUGGCUCUCUUAGGUGCUUUCCCCACCUGCAGCACGCCCCAUUCCAUCGCCAGUUGAAGUACAAAAUACAAAA ......((.(.((((((((((((...(((((((((((.....))))).))))))))))(((((.......)))))))))))))).))................................. (-25.26 = -26.54 + 1.28)

| Location | 12,117,973 – 12,118,084 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.47 |
| Mean single sequence MFE | -33.86 |
| Consensus MFE | -26.93 |
| Energy contribution | -27.33 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12117973 111 + 22407834 ACCAAACCCUGGGCC---ACUCUAAGCCAAAGU------CGGGCCUUGGGUUAUUUAUCAGAGCUGUCAGCGGGCCAUAAAUUCCGACUUUUGUCACUACCAUUUAUGGCCUAAAGAGUU ....(((((.(((((---...((.......)).------..))))).))))).....((...((.....))(((((((((((...(((....)))......)))))))))))...))... ( -35.80) >DroSec_CAF1 20113 111 + 1 UCCAAACCCUGGGCC---ACUUUAAGCCAAAGU------CGGGCCUGAGGUUAUUUAUCAGAACUGUCAGCGGGCCAUAAAUUCCGACUUUUGUCACUACCAUUUAUGGCCUAAAGAGGU .((.((((..(((((---(((((.....)))))------..)))))..))))...................(((((((((((...(((....)))......))))))))))).....)). ( -33.80) >DroSim_CAF1 19994 111 + 1 UCCAAACCCUGGGCC---ACUUUAAACCAAAGU------CGGGCCUGAGGUUAUUUAUCAGAGCUGUCAGCGGGCCAUAAAUUCCGACUUUUGUCACUACCAUUUAUGGCCUAAAGAGGU .((.((((..(((((---(((((.....)))))------..)))))..)))).....((...((.....))(((((((((((...(((....)))......)))))))))))...)))). ( -35.50) >DroEre_CAF1 20376 114 + 1 UCCAAACAACAGGCCACUACUCUAAGCCAAAGU------CGGGCCUUGGGUUAUUUAUCAGAGCUGUCAGCGGGCCAUAAAUUCCGACUUUUGUCACUACCAUUUAUGGCCUAAAGAGGU .((.(((...(((((.(.(((.........)))------.))))))...))).....((...((.....))(((((((((((...(((....)))......)))))))))))...)))). ( -30.00) >DroYak_CAF1 20302 118 + 1 UCCAAACAACAGGCCA--ACUGUAAGCCAAAGUGGGGCUCGGGCCUUGGGUUAUUUAUCAGAGCUGUCAGCGGGCCAUAAAUUCCGACUUUUGUCACUACCAUUUAUGGCCUAAAGAGGU .((.....((((....--.))))..(((((((((((((((((((.((((........)))).))).....)))))).........(((....)))....)))))).)))).......)). ( -34.20) >consensus UCCAAACCCUGGGCC___ACUCUAAGCCAAAGU______CGGGCCUUGGGUUAUUUAUCAGAGCUGUCAGCGGGCCAUAAAUUCCGACUUUUGUCACUACCAUUUAUGGCCUAAAGAGGU .((.((((..(((((..........((....))........)))))..)))).....((...((.....))(((((((((((...(((....)))......)))))))))))...)))). (-26.93 = -27.33 + 0.40)

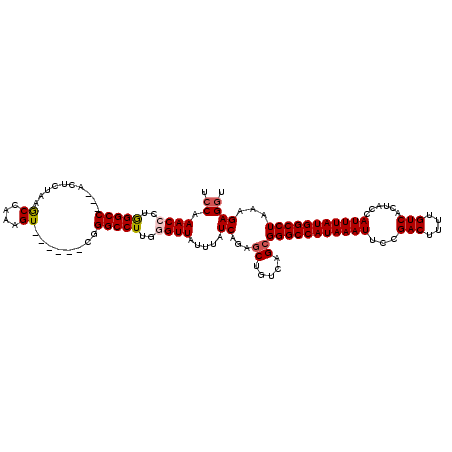
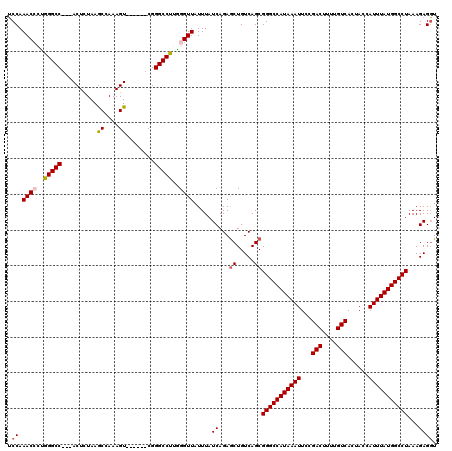
| Location | 12,118,004 – 12,118,124 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.59 |
| Mean single sequence MFE | -31.52 |
| Consensus MFE | -28.54 |
| Energy contribution | -28.94 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966414 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12118004 120 + 22407834 GGGCCUUGGGUUAUUUAUCAGAGCUGUCAGCGGGCCAUAAAUUCCGACUUUUGUCACUACCAUUUAUGGCCUAAAGAGUUAACCAAUAAAAAAAAAAAACACAAUUGGCUGGCGCAAUGC .(((.((((........)))).)))(((((((((((((((((...(((....)))......)))))))))))...........((((................))))))))))....... ( -30.49) >DroSec_CAF1 20144 118 + 1 GGGCCUGAGGUUAUUUAUCAGAACUGUCAGCGGGCCAUAAAUUCCGACUUUUGUCACUACCAUUUAUGGCCUAAAGAGGUAACCAAUAAAAC--AAAAACACAAUUGGCUGGCGCAAUGC ((..((((.........))))..))(((((((((((((((((...(((....)))......))))))))))).....((...))........--.............))))))....... ( -30.70) >DroSim_CAF1 20025 118 + 1 GGGCCUGAGGUUAUUUAUCAGAGCUGUCAGCGGGCCAUAAAUUCCGACUUUUGUCACUACCAUUUAUGGCCUAAAGAGGUAACCAAUAAAAA--AAAAACACAAUUGGCUGGCGCAAUGC .(((((((.........)))).)))(((((((((((((((((...(((....)))......))))))))))).....((...))........--.............))))))....... ( -33.20) >DroEre_CAF1 20410 114 + 1 GGGCCUUGGGUUAUUUAUCAGAGCUGUCAGCGGGCCAUAAAUUCCGACUUUUGUCACUACCAUUUAUGGCCUAAAGAGGUAACCAAUAAA------AAGUACAAUUGGCUGGCGCAAUGC (.(((...((((((((......((.....))(((((((((((...(((....)))......)))))))))))....))))))))......------.(((.......)))))).)..... ( -32.20) >DroYak_CAF1 20340 114 + 1 GGGCCUUGGGUUAUUUAUCAGAGCUGUCAGCGGGCCAUAAAUUCCGACUUUUGUCACUACCAUUUAUGGCCUAAAGAGGUAACCAAUAAA------AAAUACAAUUGGCUGGCGCAAUGC .(((.((((........)))).)))(((((((((((((((((...(((....)))......)))))))))))...........((((...------.......))))))))))....... ( -31.00) >consensus GGGCCUUGGGUUAUUUAUCAGAGCUGUCAGCGGGCCAUAAAUUCCGACUUUUGUCACUACCAUUUAUGGCCUAAAGAGGUAACCAAUAAAA___AAAAACACAAUUGGCUGGCGCAAUGC (.(((...((((((((......((.....))(((((((((((...(((....)))......)))))))))))....)))))))).................(....)...))).)..... (-28.54 = -28.94 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:17 2006