| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,248,647 – 1,248,753 |
| Length | 106 |
| Max. P | 0.954749 |

| Location | 1,248,647 – 1,248,753 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 65.92 |
| Mean single sequence MFE | -19.64 |
| Consensus MFE | -7.67 |
| Energy contribution | -7.70 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.39 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
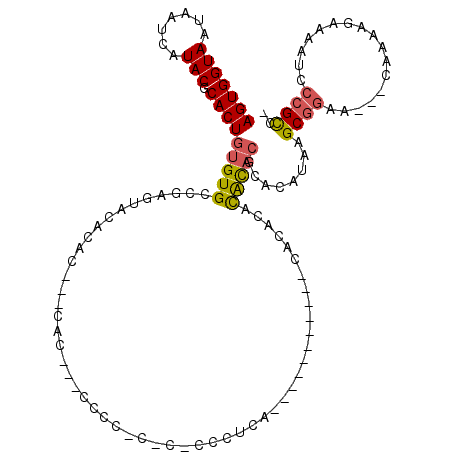
>2L_DroMel_CAF1 1248647 106 - 22407834 AGUGGUAAUAAUCAUACGCACUGUGUGCCGAGUACACACGC-CACCCCCCCCCCCCCCCCUCACA---------CACACACACACACGCAUAAGCGGAA---CAAAAGAAAAUCCCGCC- .((((............((..(((((((...))))))).))-.......................---------............(((....)))...---............)))).- ( -15.90) >DroSec_CAF1 8495 86 - 1 AGUGGUAAUAAUCAUACGCACUGUGUGGC--------------A----CCCCACCC-CCCUCA-----------CACACACACACGCAUAUAAGCGGAA---CAAAAGAAAAUCCCGCC- .(((((....)))))..((..((((((..--------------.----........-......-----------..))))))...))......((((..---............)))).- ( -14.85) >DroSim_CAF1 8959 89 - 1 AGUGGUAAUAAUCAUACGCACUGUGUGCC--------------A----CCCCACCCUCCCUCACA---------CACACACACACGCACAUAAGCGGAA---CAAAAGAAAAUCCCGCC- .((((((.......))).)))(((((((.--------------.----.................---------...........))))))).((((..---............)))).- ( -13.65) >DroEre_CAF1 21621 87 - 1 AGUGGUAAUAAUCAUACGCACUGUGUGCCGAGUACACACACACACA---------C-ACAU-----------------AGUGCA--CACAUAGGCGGAA---CAAAAGAAAAUCCCGCC- .((((((.......)))((((((((((..(.((........)).).---------)-))))-----------------))))).--)))...(((((..---............)))))- ( -25.64) >DroYak_CAF1 9039 88 - 1 AGUGGUAAUAAUCAUACGCACUGUGUGCCGAGCACACACAC-CACA---------C-CCAU-----------------CACACCCCUACAAAAGCGGAA---CAAAAGAAAAUCCCGCC- .(((((...............(((((((...))))))).))-))).---------.-....-----------------...............((((..---............)))).- ( -17.43) >DroPer_CAF1 12501 115 - 1 AGUGGUAAUAAUCAUACGCACUGUGUGACGACUACACGC---CAUGCGAGAGUA-C-GGCUCGUGCGAGCAUAAAAAGGAUAUACGAACAAAGGCCAAAGGGAAAGAAAAAAUCCCGUCU ..((((.....((...(((((.(((((.....)))))((---(.(((....)))-.-)))..)))))..........(......)))......))))..((((.........)))).... ( -30.40) >consensus AGUGGUAAUAAUCAUACGCACUGUGUGCCGAGUACACAC___CAC___CCCC_C_C_CCCUCA___________CACACACACACGCACAUAAGCGGAA___CAAAAGAAAAUCCCGCC_ (((((((.......))).))))(((((.....................................................)))))........((((.................)))).. ( -7.67 = -7.70 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:52 2006