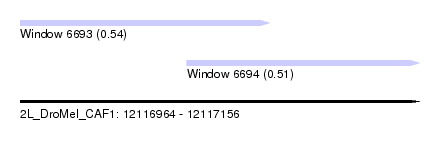
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,116,964 – 12,117,156 |
| Length | 192 |
| Max. P | 0.543934 |

| Location | 12,116,964 – 12,117,084 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.17 |
| Mean single sequence MFE | -29.07 |
| Consensus MFE | -21.26 |
| Energy contribution | -21.78 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12116964 120 + 22407834 CGAGUUAAUGAGCUUACAAAGCCACUUGAUAUCUUUAAGGUAUCAAAAGAACUUCCUGACUGCCCUUUGGUCAUCUCGAUAAUCUGACCAAAGUGUGGUGUGCCACAAAUUCGAUUAUCA ((((((..((.((.......(((((((((((((.....))))))))..................(((((((((...........))))))))).)))))..))))..))))))....... ( -32.00) >DroSec_CAF1 19096 120 + 1 CGAGUUAAUAAGCUAACAAAGUCACUUGAUAUCUUUAAGCUAUCAAAAGAACUUCCUGACUGCCCUUUGGUCAUCUCGAUAAUCUGGCCAAAGUGUGGUGUGCCACAAAUUCGAUUAUCA ((((((.............((((..((((((.(.....).)))))).((......))))))...(((((((((...........)))))))))(((((....)))))))))))....... ( -27.30) >DroSim_CAF1 18982 120 + 1 CGAGUUAAUGAGCUUACAAAGUCACUUGAUAUCUUUAAGGUAUCAAAAGAACUUCCUGACUGCCCUUUGGUCAUCUCGAUAAUCUGGCCAAAGUGUGGUGUGCCACAAAUUCGAUUAUCA ((((((..((.((.....(((((..((((((((.....))))))))..).))))....((..(.(((((((((...........))))))))).)..))..))))..))))))....... ( -32.30) >DroEre_CAF1 19380 112 + 1 --------UGAUCUUACAAAGUUACUCGAUAUUGUAAAGGUAUCAAAUGAACUUCCUGAAUGCCCUUUGGUCAUCUCGAUAAUCUUGCCAAAGUGUGGUGUGCCACAAAUUCGAUUAUCA --------(((((((((((.(((....))).)))))).(((((((...........)).)))))....)))))....(((((((.........(((((....))))).....))))))). ( -24.14) >DroYak_CAF1 19305 120 + 1 CGAGUUAAUGGGCUUACAACGUCACUCGAUAUCUUUAAGGUAUCAAAUGAACUUCCUGAAUGCCCUUCGGUCAUCUCGAUAAUCUGGCCAAAGUGUGGUGUGCCACAAAUUCGAUUAUCA ((((((...((((...((...(((...((((((.....))))))...)))......))...))))...(((((...........)))))....(((((....)))))))))))....... ( -29.60) >consensus CGAGUUAAUGAGCUUACAAAGUCACUUGAUAUCUUUAAGGUAUCAAAAGAACUUCCUGACUGCCCUUUGGUCAUCUCGAUAAUCUGGCCAAAGUGUGGUGUGCCACAAAUUCGAUUAUCA ...................((((....((((((.....))))))....(((.............(((((((((...........)))))))))(((((....)))))..))))))).... (-21.26 = -21.78 + 0.52)

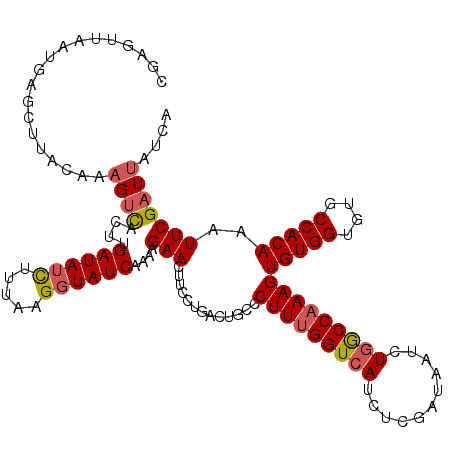
| Location | 12,117,044 – 12,117,156 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.93 |
| Mean single sequence MFE | -33.80 |
| Consensus MFE | -28.66 |
| Energy contribution | -29.14 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.85 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12117044 112 + 22407834 AAUCUGACCAAAGUGUGGUGUGCCACAAAUUCGAUUAUCAGGGCCGGGAGCUG---AGCUUCGACCGCCAUCUCAAGGUCGAAAAAAAAAGUAUCUUGUAUCU-GGUAUCUGGUAU---- ......((((...(((((....))))).....(((.((((((..((((((((.---...(((((((..........)))))))......))).)))))..)))-))))))))))..---- ( -34.50) >DroSec_CAF1 19176 114 + 1 AAUCUGGCCAAAGUGUGGUGUGCCACAAAUUCGAUUAUCAGGGCCGGGAGCUGCUAAGCUUCGACCGCCAUCUCAAGGUCGAAAA--AAAGUAUCUUGUAUCUUGGUAUCCGGUAU---- ..(((((((....(((((....))))).....(.....)..))))))).(((....)))(((((((..........)))))))..--...(((((..((((....))))..)))))---- ( -34.20) >DroSim_CAF1 19062 114 + 1 AAUCUGGCCAAAGUGUGGUGUGCCACAAAUUCGAUUAUCAGGGCCGGGAGCUGCUAAGCUUCGACCGCCAUCUCAAGGUCGAAAA--AAAGUAUCUUGUAUCUGGGUAUCUGGUAU---- ..(((((((....(((((....))))).....(.....)..))))))).(((....)))(((((((..........)))))))..--...(((((..((((....))))..)))))---- ( -35.10) >DroEre_CAF1 19452 113 + 1 AAUCUUGCCAAAGUGUGGUGUGCCACAAAUUCGAUUAUCAGGGCCGGGAGCUG---AGCUUCGACCGCCAUCUCAAGGUCGGAA---AAAGUAUCUUGUGUCUU-GUAUCUUGUAUCUGG .......(((...(((((....))))).....(((...((((((((((((((.---...(((((((..........))))))).---..))).))))).)))))-).))).......))) ( -33.10) >DroYak_CAF1 19385 109 + 1 AAUCUGGCCAAAGUGUGGUGUGCCACAAAUUCGAUUAUCAGGGCCGGGAGCUG---AGCCUCGACCGCCAUCUCAAGGUCGAAA---AAAGUAUCUCGUAUCUU-GUAUCUGGUAU---- ......((((...(((((....))))).....(((...(((((.((((((((.---....((((((..........))))))..---..))).)))))..))))-).)))))))..---- ( -32.10) >consensus AAUCUGGCCAAAGUGUGGUGUGCCACAAAUUCGAUUAUCAGGGCCGGGAGCUG___AGCUUCGACCGCCAUCUCAAGGUCGAAAA__AAAGUAUCUUGUAUCUUGGUAUCUGGUAU____ ..(((((((....(((((....))))).....(.....)..))))))).(((....)))(((((((..........))))))).......(((((..((((....))))..))))).... (-28.66 = -29.14 + 0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:11 2006