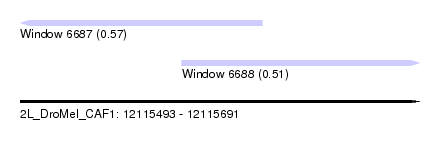
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,115,493 – 12,115,691 |
| Length | 198 |
| Max. P | 0.567758 |

| Location | 12,115,493 – 12,115,613 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.83 |
| Mean single sequence MFE | -26.25 |
| Consensus MFE | -19.42 |
| Energy contribution | -20.50 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12115493 120 - 22407834 AUAUUUUCUUUUCUUUUGGCUAAUAAAUAAGCAAUUUGGUGGCUGCUUCUUUUACUGUCAUCGACUAUUUCUUUAAUUGGUCUGCGCAAGCUGGUAAGCAUUAUUUAAUUAGCAUAGAUU ...........(((....((((((((((((((.....((((((.............))))))(((((..........))))).((....))......)).)))))).))))))..))).. ( -26.82) >DroSec_CAF1 17592 120 - 1 AUGUUUUCUUUUCUUUUGGCUAAUAAAUAAGCAAUUUGGUGACUGCUUCUUCUACUGUCAUCGACUAUUUCUUUAAUUGGUCUGCGCAAGCUGGUAAGCAUUAUUUAAUUAGCAUAGAUU ...........(((....((((((((((((((.....((((((.............))))))(((((..........))))).((....))......)).)))))).))))))..))).. ( -27.42) >DroSim_CAF1 17427 120 - 1 AUAUUUUCUUUUCUUUUGGCUAAUAAAUAAGCAAUUUGCUGGCUGCUUCUUCUACUGUCAUCGACUAUUUCUUUAAUUGGUCUGCGCAGGCUGGUAAGCAUUAUUUAAUUAGCAUAGAUU ...........(((....((((((((((((((...((((..(((((..........(....)(((((..........)))))...))).))..)))))).)))))).))))))..))).. ( -26.40) >DroEre_CAF1 17863 120 - 1 AUAUUUCAUUUUCUUUUGGCUAAUAAAUAAGCAAUUUAGUAGCUGCUUCUUCUGCUGUCAUCGAAUACUUCUUUAAUUGGCCUGCGCAAGCUGGUAAGCAUUAUUUAAUUAGCAUAGAUG ...........(((....((((((((((((((.(((..((..(.((.......)).)..))..))).............(((.((....)).)))..)).)))))).))))))..))).. ( -26.80) >DroYak_CAF1 17765 120 - 1 AUAUUUUCCCUUCUUUUGGCUAAUAAAUAAGCAAUUUGGUAGCUGCUUCUUCUGCUGUCAUCGACUAUUUCUUUAAUUGGUCUGCGCAAGCCGGUAAGCACUAUAUAAUUUGCAUAGAUG .((((..((........))..)))).....((((....((((.((((.....(((((.....(((((..........))))).((....))))))))))))))).....))))....... ( -23.80) >consensus AUAUUUUCUUUUCUUUUGGCUAAUAAAUAAGCAAUUUGGUGGCUGCUUCUUCUACUGUCAUCGACUAUUUCUUUAAUUGGUCUGCGCAAGCUGGUAAGCAUUAUUUAAUUAGCAUAGAUU ...........(((....((((((((((((((...((((((((.............))))))))................((.((....)).))...)).)))))).))))))..))).. (-19.42 = -20.50 + 1.08)

| Location | 12,115,573 – 12,115,691 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.29 |
| Mean single sequence MFE | -21.04 |
| Consensus MFE | -19.96 |
| Energy contribution | -19.44 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.95 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507778 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12115573 118 + 22407834 ACCAAAUUGCUUAUUUAUUAGCCAAAAGAAAAGAAAAUAUCUGGAAACGGCAUGCUAAUGAUGUAAACUAAGCAUUUAACUGUUAAGAUGCCAGCAGCGGCAGGCAAACAU-A-AAAAGU .......((((((.......(((........(((.....)))(....)))).(((.......)))...))))))......((((....((((.((....)).)))))))).-.-...... ( -20.80) >DroSec_CAF1 17672 118 + 1 ACCAAAUUGCUUAUUUAUUAGCCAAAAGAAAAGAAAACAUCUGGAAACGGCAUGCUAAUGAUGUAAACUAAGCAUUUAACUGUUAAGAUGCCAGCAGCGGCAGGCAAACAU-A-AAAAGU .......((((((.......(((........(((.....)))(....)))).(((.......)))...))))))......((((....((((.((....)).)))))))).-.-...... ( -20.80) >DroSim_CAF1 17507 118 + 1 AGCAAAUUGCUUAUUUAUUAGCCAAAAGAAAAGAAAAUAUCUGGAAACGGCAUGCUAAUGAUGUAAACUAAGCAUUUAACUGUUAAGAUGCCAGCAGCGGCAGGCAAACAU-A-AAAAGU ((((((.((((((.......(((........(((.....)))(....)))).(((.......)))...)))))).))...))))....((((.((....)).)))).....-.-...... ( -21.20) >DroEre_CAF1 17943 119 + 1 ACUAAAUUGCUUAUUUAUUAGCCAAAAGAAAAUGAAAUAUCUGGAAACGGCAUGCUAAUGAUGUAAACUAAGCAUUUAACUGUUAAGAUGCCAGCAGCGGCAAGCAAACAA-AAAGAAGU ......((((((..((((((((((........))......(((....)))...))))))))......((.((((......)))).))..(((......)))))))))....-........ ( -21.70) >DroYak_CAF1 17845 120 + 1 ACCAAAUUGCUUAUUUAUUAGCCAAAAGAAGGGAAAAUAUCUGGAAACGGCAUGCUAAUGAUGUAAACUAAGCAUUUAACUGUUAAGAUGCCAGCAGCAGCAAGCAAACGAGAAAGAAGC ......((((((........(((........(((.....)))(....)))).((((..((..(....)...((((((........))))))))..))))..))))))............. ( -20.70) >consensus ACCAAAUUGCUUAUUUAUUAGCCAAAAGAAAAGAAAAUAUCUGGAAACGGCAUGCUAAUGAUGUAAACUAAGCAUUUAACUGUUAAGAUGCCAGCAGCGGCAGGCAAACAU_A_AAAAGU ......((((((........(((........(((.....)))(....))))..(((..((.((........((((((........)))))))).))..)))))))))............. (-19.96 = -19.44 + -0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:05 2006