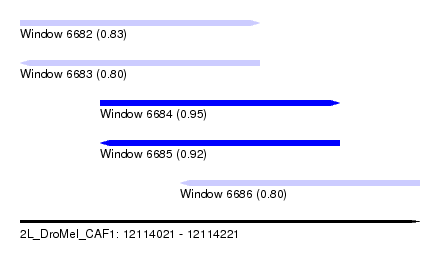
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,114,021 – 12,114,221 |
| Length | 200 |
| Max. P | 0.953247 |

| Location | 12,114,021 – 12,114,141 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.80 |
| Mean single sequence MFE | -25.46 |
| Consensus MFE | -22.98 |
| Energy contribution | -22.86 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12114021 120 + 22407834 GCCCGUCCGAACUUUUAUCUUUGCAAUCGUUCCGCGAACUUUGGCCAAUCUUAUGGACAAAAUCUAAAGCGAAAUCUAAUUUUUAAUUGGCCAACAAACUGCGGACAAGCGACAUUUCAU ((..(((((..........(((((.........)))))..(((((((((....((((.....))))....((((......)))).))))))))).......)))))..)).......... ( -25.30) >DroSec_CAF1 16130 113 + 1 ------ACG-ACAUUUAUCUUUGCAAUCGUUCCGCGAACUUUGGCCAAUCUCAUGGACAAAAUCUAAUGCGAAAUCUAAUUUUUAAUUGGCCAACAAACUGCGGACAAGCGACAUUUCAU ------...-................((((((((((....(((((((((....((((.....))))....((((......)))).))))))))).....))))))...))))........ ( -24.30) >DroSim_CAF1 15957 119 + 1 CCCCGUCCG-ACAUUUAUCUUUGCAAUCGUUCCGCGAACUUUGGCCAAUCACAUGGACAAAAUCUAAAGCGAAAUCUAAUUUUUAAUUGGCCAACAAACUGCGGACAAGCGACAUUUCAU ....(((((-.((......(((((.........)))))..(((((((((....((((.....))))....((((......)))).))))))))).....))))))).............. ( -24.30) >DroEre_CAF1 16375 119 + 1 ACCCAUCAG-ACAUUUAUCUUUGCAAUCGUUCCGCGAGCUUUGGCCAAUCUCAUGGGCAAAAUCUGAAGCGAAAUCUAAUUUUUAAUUGGCCAACAAACUGCGGACAAGGGACAUUUCAU .(((...((-(......)))........((.(((((....(((((((((.(((.((......)))))...((((......)))).))))))))).....)))))))..)))......... ( -25.50) >DroYak_CAF1 16254 119 + 1 CCCCGUCAG-GCAUUUAUCUUUGCAAUCGUUCCGCGAACUUUGGCCAAUCUCGUGGACAAAAUCUAAAGCGAAAUCUAAUUUUUAAUUGGCCAACAAACUGCGGACAAGCGACAUUUCAU ....(((..-(((........)))....((.(((((....(((((((((....((((.....))))....((((......)))).))))))))).....)))))))....)))....... ( -27.90) >consensus CCCCGUCCG_ACAUUUAUCUUUGCAAUCGUUCCGCGAACUUUGGCCAAUCUCAUGGACAAAAUCUAAAGCGAAAUCUAAUUUUUAAUUGGCCAACAAACUGCGGACAAGCGACAUUUCAU ..........................((((((((((....(((((((((....((((.....))))....((((......)))).))))))))).....))))))...))))........ (-22.98 = -22.86 + -0.12)

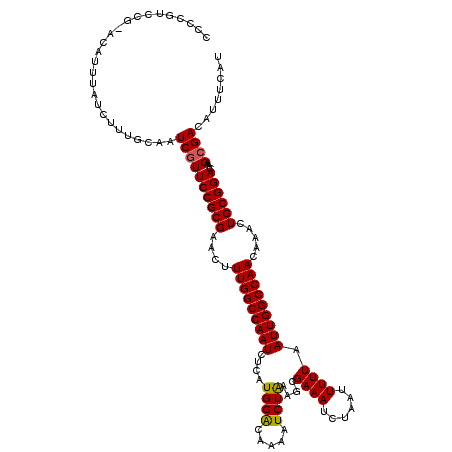
| Location | 12,114,021 – 12,114,141 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.80 |
| Mean single sequence MFE | -32.82 |
| Consensus MFE | -27.13 |
| Energy contribution | -27.33 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12114021 120 - 22407834 AUGAAAUGUCGCUUGUCCGCAGUUUGUUGGCCAAUUAAAAAUUAGAUUUCGCUUUAGAUUUUGUCCAUAAGAUUGGCCAAAGUUCGCGGAACGAUUGCAAAGAUAAAAGUUCGGACGGGC ..........((((((((((((((..((((((((((........((..((......))..))........)))))))))).((((...))))))))))...((.......)))))))))) ( -35.79) >DroSec_CAF1 16130 113 - 1 AUGAAAUGUCGCUUGUCCGCAGUUUGUUGGCCAAUUAAAAAUUAGAUUUCGCAUUAGAUUUUGUCCAUGAGAUUGGCCAAAGUUCGCGGAACGAUUGCAAAGAUAAAUGU-CGU------ .((....((((....(((((.(....((((((((((........((..((......))..))........))))))))))....)))))).))))..))..(((....))-)..------ ( -28.59) >DroSim_CAF1 15957 119 - 1 AUGAAAUGUCGCUUGUCCGCAGUUUGUUGGCCAAUUAAAAAUUAGAUUUCGCUUUAGAUUUUGUCCAUGUGAUUGGCCAAAGUUCGCGGAACGAUUGCAAAGAUAAAUGU-CGGACGGGG ...........(((((((((((((..(((((((((((.......((..((......))..)).......))))))))))).((((...))))))))))...(((....))-)))))))). ( -36.94) >DroEre_CAF1 16375 119 - 1 AUGAAAUGUCCCUUGUCCGCAGUUUGUUGGCCAAUUAAAAAUUAGAUUUCGCUUCAGAUUUUGCCCAUGAGAUUGGCCAAAGCUCGCGGAACGAUUGCAAAGAUAAAUGU-CUGAUGGGU .........((((((((((((((...((((((((((........((..((......))..))........)))))))))).))).)))).))))......((((....))-))...))). ( -32.89) >DroYak_CAF1 16254 119 - 1 AUGAAAUGUCGCUUGUCCGCAGUUUGUUGGCCAAUUAAAAAUUAGAUUUCGCUUUAGAUUUUGUCCACGAGAUUGGCCAAAGUUCGCGGAACGAUUGCAAAGAUAAAUGC-CUGACGGGG ......((((..((((((((.(....((((((((((........((..((......))..))........))))))))))....))))).))))..(((........)))-..))))... ( -29.89) >consensus AUGAAAUGUCGCUUGUCCGCAGUUUGUUGGCCAAUUAAAAAUUAGAUUUCGCUUUAGAUUUUGUCCAUGAGAUUGGCCAAAGUUCGCGGAACGAUUGCAAAGAUAAAUGU_CGGACGGGG .((....((((....(((((.(....((((((((((........((..((......))..))........))))))))))....)))))).))))..))..................... (-27.13 = -27.33 + 0.20)

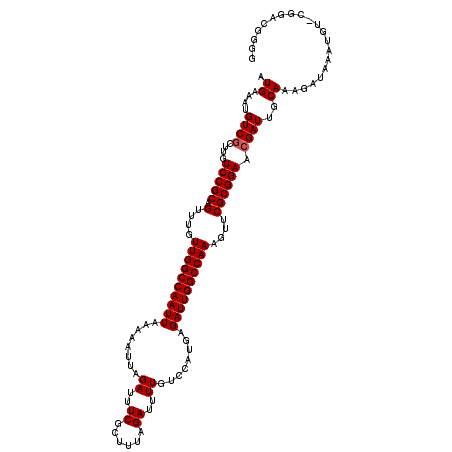
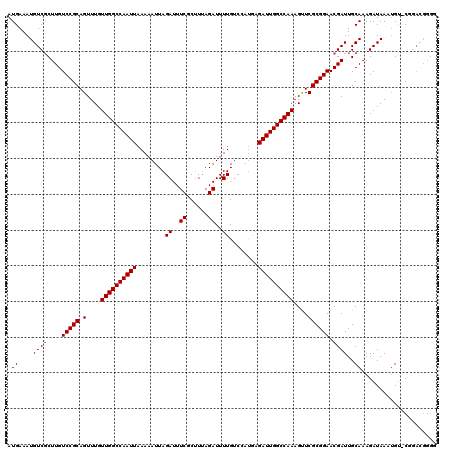
| Location | 12,114,061 – 12,114,181 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -26.80 |
| Consensus MFE | -25.06 |
| Energy contribution | -24.78 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12114061 120 + 22407834 UUGGCCAAUCUUAUGGACAAAAUCUAAAGCGAAAUCUAAUUUUUAAUUGGCCAACAAACUGCGGACAAGCGACAUUUCAUGCUUAUCGGCAGUACAACCAGCAAUUAAUCUCCCUUCGGU (((((((((....((((.....))))....((((......)))).)))))))))...(((((.((.(((((........))))).)).)))))....................(....). ( -26.80) >DroSec_CAF1 16163 120 + 1 UUGGCCAAUCUCAUGGACAAAAUCUAAUGCGAAAUCUAAUUUUUAAUUGGCCAACAAACUGCGGACAAGCGACAUUUCAUGCUUAUCGGCGGUACAACCAGCAAUUAAUCUCCCUUCGGU (((((((((....((((.....))))....((((......)))).)))))))))...(((((.((.(((((........))))).)).)))))....................(....). ( -26.10) >DroSim_CAF1 15996 120 + 1 UUGGCCAAUCACAUGGACAAAAUCUAAAGCGAAAUCUAAUUUUUAAUUGGCCAACAAACUGCGGACAAGCGACAUUUCAUGCUUAUCGGCAGUACAACCAGCAAUUAAUCUCCCUUCGGU (((((((((....((((.....))))....((((......)))).)))))))))...(((((.((.(((((........))))).)).)))))....................(....). ( -26.80) >DroEre_CAF1 16414 120 + 1 UUGGCCAAUCUCAUGGGCAAAAUCUGAAGCGAAAUCUAAUUUUUAAUUGGCCAACAAACUGCGGACAAGGGACAUUUCAUGCUUAUCUGCAGUACAACCAGCAAUUAAUCUAUCUUCGGU (((.(((......))).)))...((((((.(....)......(((((((........((((((((.(((.((....))...))).))))))))........))))))).....)))))). ( -25.39) >DroYak_CAF1 16293 120 + 1 UUGGCCAAUCUCGUGGACAAAAUCUAAAGCGAAAUCUAAUUUUUAAUUGGCCAACAAACUGCGGACAAGCGACAUUUCAUGCUUAUCGGCAGUACAACCAGCAAUUAAUCGCCCUUCGGC (((((((((....((((.....))))....((((......)))).)))))))))...(((((.((.(((((........))))).)).))))).................(((....))) ( -28.90) >consensus UUGGCCAAUCUCAUGGACAAAAUCUAAAGCGAAAUCUAAUUUUUAAUUGGCCAACAAACUGCGGACAAGCGACAUUUCAUGCUUAUCGGCAGUACAACCAGCAAUUAAUCUCCCUUCGGU (((((((((....((((.....))))....((((......)))).)))))))))...(((((.((.(((((........))))).)).)))))........................... (-25.06 = -24.78 + -0.28)


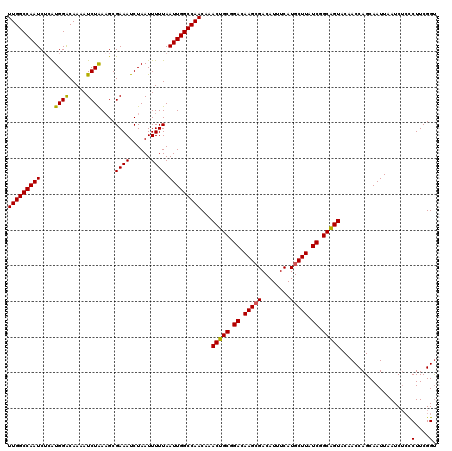
| Location | 12,114,061 – 12,114,181 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -34.02 |
| Consensus MFE | -28.99 |
| Energy contribution | -29.07 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12114061 120 - 22407834 ACCGAAGGGAGAUUAAUUGCUGGUUGUACUGCCGAUAAGCAUGAAAUGUCGCUUGUCCGCAGUUUGUUGGCCAAUUAAAAAUUAGAUUUCGCUUUAGAUUUUGUCCAUAAGAUUGGCCAA .((....)).(((.((((((.(((......)))(((((((..........))))))).)))))).)))((((((((........((..((......))..))........)))))))).. ( -34.09) >DroSec_CAF1 16163 120 - 1 ACCGAAGGGAGAUUAAUUGCUGGUUGUACCGCCGAUAAGCAUGAAAUGUCGCUUGUCCGCAGUUUGUUGGCCAAUUAAAAAUUAGAUUUCGCAUUAGAUUUUGUCCAUGAGAUUGGCCAA .((....)).(((.((((((.(((......)))(((((((..........))))))).)))))).)))((((((((........((..((......))..))........)))))))).. ( -33.39) >DroSim_CAF1 15996 120 - 1 ACCGAAGGGAGAUUAAUUGCUGGUUGUACUGCCGAUAAGCAUGAAAUGUCGCUUGUCCGCAGUUUGUUGGCCAAUUAAAAAUUAGAUUUCGCUUUAGAUUUUGUCCAUGUGAUUGGCCAA .((....)).(((.((((((.(((......)))(((((((..........))))))).)))))).)))(((((((((.......((..((......))..)).......))))))))).. ( -36.34) >DroEre_CAF1 16414 120 - 1 ACCGAAGAUAGAUUAAUUGCUGGUUGUACUGCAGAUAAGCAUGAAAUGUCCCUUGUCCGCAGUUUGUUGGCCAAUUAAAAAUUAGAUUUCGCUUCAGAUUUUGCCCAUGAGAUUGGCCAA ((((..(((......)))..))))...(((((.((((((...((....)).)))))).)))))...((((((((((........((..((......))..))........)))))))))) ( -28.49) >DroYak_CAF1 16293 120 - 1 GCCGAAGGGCGAUUAAUUGCUGGUUGUACUGCCGAUAAGCAUGAAAUGUCGCUUGUCCGCAGUUUGUUGGCCAAUUAAAAAUUAGAUUUCGCUUUAGAUUUUGUCCACGAGAUUGGCCAA (((..((((((((((((((((..(...(((((.(((((((..........))))))).)))))..)..)).)))))))..........))))))).(((((((....))))))))))... ( -37.80) >consensus ACCGAAGGGAGAUUAAUUGCUGGUUGUACUGCCGAUAAGCAUGAAAUGUCGCUUGUCCGCAGUUUGUUGGCCAAUUAAAAAUUAGAUUUCGCUUUAGAUUUUGUCCAUGAGAUUGGCCAA ((((..((........))..))))...(((((.(((((((..........))))))).)))))...((((((((((........((..((......))..))........)))))))))) (-28.99 = -29.07 + 0.08)

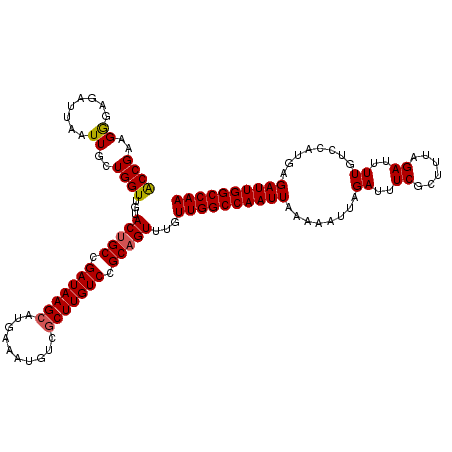
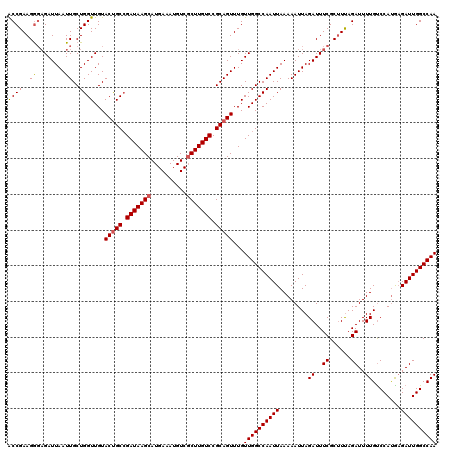
| Location | 12,114,101 – 12,114,221 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.72 |
| Mean single sequence MFE | -34.12 |
| Consensus MFE | -27.66 |
| Energy contribution | -28.66 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12114101 120 - 22407834 AGAGAAAGAGAGCAUCUGCUAAGAGAGUCUUCUCCAUUCCACCGAAGGGAGAUUAAUUGCUGGUUGUACUGCCGAUAAGCAUGAAAUGUCGCUUGUCCGCAGUUUGUUGGCCAAUUAAAA ..(((.....(((....))).......)))(((((.(((....))).)))))(((((((((..(...(((((.(((((((..........))))))).)))))..)..)).))))))).. ( -35.30) >DroSec_CAF1 16203 120 - 1 AGAGAAAGAGAGUAUCUGCUAAGAGAGUCUUCUCCACUCCACCGAAGGGAGAUUAAUUGCUGGUUGUACCGCCGAUAAGCAUGAAAUGUCGCUUGUCCGCAGUUUGUUGGCCAAUUAAAA .((((...(((.(.(((....))).).)))))))..((((.......)))).(((((((..((.....))((((((((((..((....))((......)).))))))))))))))))).. ( -31.70) >DroSim_CAF1 16036 120 - 1 AGAGAAAGAGAGAAUCUGCUAAGAGAGUCUUCUCCACUCCACCGAAGGGAGAUUAAUUGCUGGUUGUACUGCCGAUAAGCAUGAAAUGUCGCUUGUCCGCAGUUUGUUGGCCAAUUAAAA .(((...((((((.(((....)))...)).))))..)))..((....))...(((((((((..(...(((((.(((((((..........))))))).)))))..)..)).))))))).. ( -36.60) >DroEre_CAF1 16454 115 - 1 AGAGAAAGAGAGAAUCCGCUAAGAGAGUCUUCUC-----UACCGAAGAUAGAUUAAUUGCUGGUUGUACUGCAGAUAAGCAUGAAAUGUCCCUUGUCCGCAGUUUGUUGGCCAAUUAAAA ......(((((((.(((.....).)).)).))))-----)............(((((((((..(...(((((.((((((...((....)).)))))).)))))..)..)).))))))).. ( -27.00) >DroYak_CAF1 16333 115 - 1 AGAGAAAGAGAGAAUCUGCUAAGAGAGUCUUCUC-----CGCCGAAGGGCGAUUAAUUGCUGGUUGUACUGCCGAUAAGCAUGAAAUGUCGCUUGUCCGCAGUUUGUUGGCCAAUUAAAA .......((((((.(((....)))...)).))))-----((((....)))).(((((((((..(...(((((.(((((((..........))))))).)))))..)..)).))))))).. ( -40.00) >consensus AGAGAAAGAGAGAAUCUGCUAAGAGAGUCUUCUCCA_UCCACCGAAGGGAGAUUAAUUGCUGGUUGUACUGCCGAUAAGCAUGAAAUGUCGCUUGUCCGCAGUUUGUUGGCCAAUUAAAA .(((((.((.....(((....)))...))))))).......((....))...(((((((((..(...(((((.(((((((..........))))))).)))))..)..)).))))))).. (-27.66 = -28.66 + 1.00)

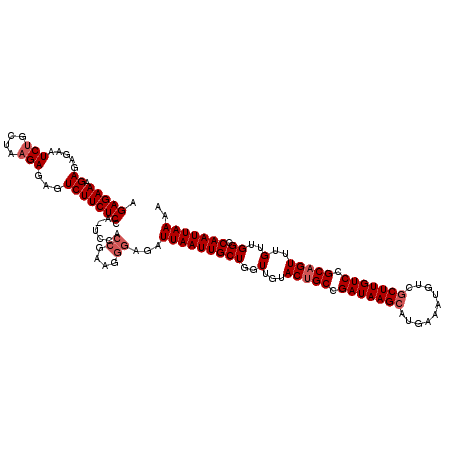
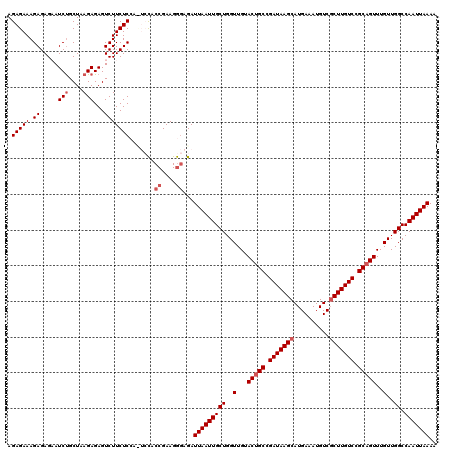
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:03 2006