| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,091,900 – 12,092,032 |
| Length | 132 |
| Max. P | 0.863800 |

| Location | 12,091,900 – 12,092,001 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.04 |
| Mean single sequence MFE | -28.70 |
| Consensus MFE | -14.62 |
| Energy contribution | -14.60 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
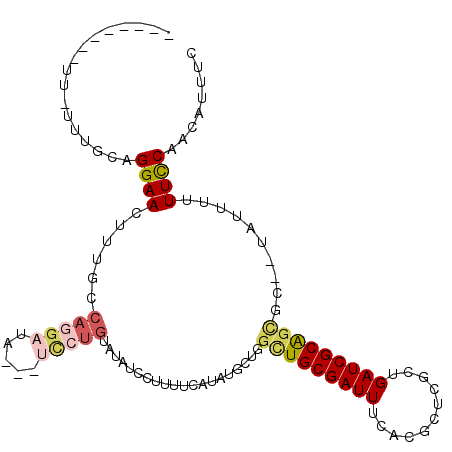
>2L_DroMel_CAF1 12091900 101 + 22407834 AAAUCGAGCA----------UCUUUCGCUGGAAGGAUGCGGAAAUGUUGAAAAAAAUA---------GCGAUGCGAUCAGCGAGCGUGAAAUCGCAGUCAGCAUAUGAAAAGGAUGUACA ..(((..(((----------((((((....)))))))))....(((((((........---------((..(((.....))).))(((....)))..)))))))........)))..... ( -28.50) >DroSec_CAF1 28301 101 + 1 AAAUCGGGCA----------UCUUUCGCUGGAAGGAUGCCGAAAUGUUGGAAAAAAUA---------GCGCUGCGAUCAGCGAGCGUGAAAUCGCAACCAGCAUAUGAAAAGGAUAUACA ..(((.((((----------((((((....))))))))))...(((((((........---------.(((((....))))).(((......)))..)))))))........)))..... ( -36.30) >DroEre_CAF1 28574 99 + 1 AAAUCGGGCA----------UCUUUCGCUGGAAGGAUACGGAAAUGUGGAAAA--AAA---------GCGCUGCGAUCAGCGAGCGUGAAAUCGCAGCCAGCAUAUGAAAAGGAUAUACA ...(((.(((----------((((((....)))))))..((...(((((....--...---------(((((((.....)).)))))....))))).)).))...)))............ ( -25.80) >DroYak_CAF1 30421 101 + 1 AAAUCGGGCA----------UCUUUCGCUGGAAGGAUACGGAAAUGUCGGAAAAAAUA---------GCGCUGCGAUCAGCGAGCGUGAAAUCGCAGCUAGCAUAUGAAAAGGAUGUACA .....(..((----------(((((.((((........(((.....))).......))---------))((((((((..((....))...))))))))...........)))))))..). ( -28.26) >DroAna_CAF1 29833 102 + 1 AAAUCGCGCC----------UCCUUCCCAGUAAGGAUGCUGAAAA-UUGAAAAAAAUAA-------AACACCGCGAUCAGCGACCGUGAAAUCGCAGCCAAUAUAUGGAUAGGAUAUGCA .....(((..----------((((..(((....((.((((((...-.............-------..........)))))).))(((....)))..........)))..))))..))). ( -23.47) >DroPer_CAF1 32399 118 + 1 AAAUUGCGCCUCUUCCACCUUCUUUCGCUGGAAGGAUGCGGAAAAGUCGGAAAAAAUAAAUAUAUAAAUGCUGCGAUCAGUGACCGUGAAAUCGCAGCCAAUAUAUGGAAAGGUUAUA-- .......((((.((((.((((((......))))))........................((((((....((((((((((.......)))..)))))))..))))))))))))))....-- ( -29.90) >consensus AAAUCGGGCA__________UCUUUCGCUGGAAGGAUGCGGAAAUGUUGAAAAAAAUA_________GCGCUGCGAUCAGCGAGCGUGAAAUCGCAGCCAGCAUAUGAAAAGGAUAUACA ..(((...............((((((....))))))...............................((((((((((..((....))...))))))))..))..........)))..... (-14.62 = -14.60 + -0.02)

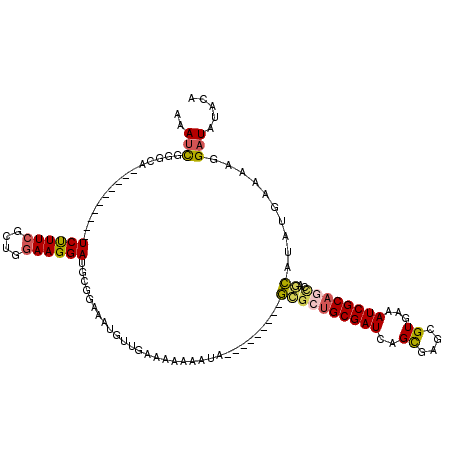

| Location | 12,091,930 – 12,092,032 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.11 |
| Mean single sequence MFE | -25.83 |
| Consensus MFE | -14.34 |
| Energy contribution | -16.45 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12091930 102 + 22407834 GAAAUGUUGAAAAAAAUA--GCGAUGCGAUCAGCGAGCGUGAAAUCGCAGUCAGCAUAUGAAAAGGAUGUACAGGAUCGUAUCCUGGCAAAGUUCCUGCAAA-AA-------- ...(((((((........--((..(((.....))).))(((....)))..))))))).((...(((((((.((((((...)))))))))....)))).))..-..-------- ( -26.30) >DroSec_CAF1 28331 102 + 1 GAAAUGUUGGAAAAAAUA--GCGCUGCGAUCAGCGAGCGUGAAAUCGCAACCAGCAUAUGAAAAGGAUAUACAGGAUCGUAUCCUGGCAAAGUUCCUGCAAA-AA-------- ...(((((((........--.(((((....))))).(((......)))..))))))).((...((((....((((((...)))))).......)))).))..-..-------- ( -30.30) >DroSim_CAF1 28707 102 + 1 GAAAUGUUGGAAAAAAUA--GCGCUGCGAUCAGCGAGCGUGAAAUCGCAGCCAGCAUAUGAAAAGGAUAUACAGGAUCGUAUCCUGGCAAAGUUCCUGCAAA-AA-------- ....(((.((((......--((((((((((..((....))...))))))))..))...((...(((((((........)))))))..))...)))).)))..-..-------- ( -30.50) >DroEre_CAF1 28604 103 + 1 GAAAUGUGGAAAA--AAA--GCGCUGCGAUCAGCGAGCGUGAAAUCGCAGCCAGCAUAUGAAAAGGAUAUACAGGC---GAUCCUGGCAAAGUUCCCGAAAA-AAUGUG--UG .............--...--(.((((((((..((....))...))))))))).((((((.....(((....((((.---...)))).......)))......-.)))))--). ( -25.00) >DroYak_CAF1 30451 107 + 1 GAAAUGUCGGAAAAAAUA--GCGCUGCGAUCAGCGAGCGUGAAAUCGCAGCUAGCAUAUGAAAAGGAUGUACAUAA---GAUCCUGGCAAAGUUCCCGAAAA-AAUGUGUGUG ......((((........--((((((((((..((....))...))))))))..))...((...(((((.(.....)---.)))))..))......))))...-.......... ( -27.20) >DroAna_CAF1 29863 96 + 1 GAAAA-UUGAAAAAAAUAAAACACCGCGAUCAGCGACCGUGAAAUCGCAGCCAAUAUAUGGAUAGGAUAUGCACAA--------------UAUUCCCAAAAAAAAUGUG--UG .....-..............((((.(((((..((....))...)))))..........(((...((((((.....)--------------))))))))........)))--). ( -15.70) >consensus GAAAUGUUGAAAAAAAUA__GCGCUGCGAUCAGCGAGCGUGAAAUCGCAGCCAGCAUAUGAAAAGGAUAUACAGGA___UAUCCUGGCAAAGUUCCCGAAAA_AA________ ...(((((((..........(((((((.....))).))))..........))))))).......(((....(((((.....))))).......)))................. (-14.34 = -16.45 + 2.11)

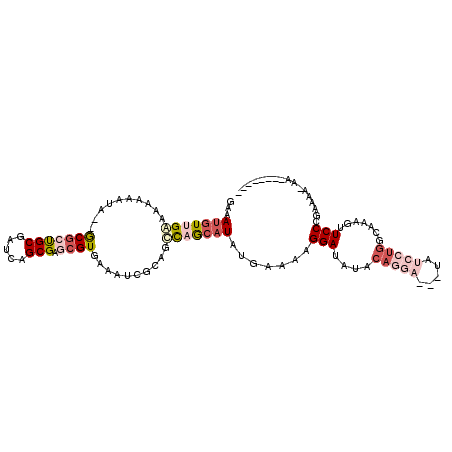
| Location | 12,091,930 – 12,092,032 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.11 |
| Mean single sequence MFE | -22.00 |
| Consensus MFE | -15.62 |
| Energy contribution | -16.40 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12091930 102 - 22407834 --------UU-UUUGCAGGAACUUUGCCAGGAUACGAUCCUGUACAUCCUUUUCAUAUGCUGACUGCGAUUUCACGCUCGCUGAUCGCAUCGC--UAUUUUUUUCAACAUUUC --------..-...((((((....(((.(((((...))))))))..))))......((((.(((.((((........)))).).)))))).))--.................. ( -18.10) >DroSec_CAF1 28331 102 - 1 --------UU-UUUGCAGGAACUUUGCCAGGAUACGAUCCUGUAUAUCCUUUUCAUAUGCUGGUUGCGAUUUCACGCUCGCUGAUCGCAGCGC--UAUUUUUUCCAACAUUUC --------..-......((((....((((((((...))))))....................(((((((((...........)))))))))))--......))))........ ( -22.30) >DroSim_CAF1 28707 102 - 1 --------UU-UUUGCAGGAACUUUGCCAGGAUACGAUCCUGUAUAUCCUUUUCAUAUGCUGGCUGCGAUUUCACGCUCGCUGAUCGCAGCGC--UAUUUUUUCCAACAUUUC --------..-......((((....((((((((...))))))....................(((((((((...........)))))))))))--......))))........ ( -24.70) >DroEre_CAF1 28604 103 - 1 CA--CACAUU-UUUUCGGGAACUUUGCCAGGAUC---GCCUGUAUAUCCUUUUCAUAUGCUGGCUGCGAUUUCACGCUCGCUGAUCGCAGCGC--UUU--UUUUCCACAUUUC ..--......-.....(((((....((((((...---.))))....................(((((((((...........)))))))))))--...--.)))))....... ( -24.80) >DroYak_CAF1 30451 107 - 1 CACACACAUU-UUUUCGGGAACUUUGCCAGGAUC---UUAUGUACAUCCUUUUCAUAUGCUAGCUGCGAUUUCACGCUCGCUGAUCGCAGCGC--UAUUUUUUCCGACAUUUC ..........-...(((((((....((.(((((.---........)))))............(((((((((...........)))))))))))--.....)))))))...... ( -25.80) >DroAna_CAF1 29863 96 - 1 CA--CACAUUUUUUUUGGGAAUA--------------UUGUGCAUAUCCUAUCCAUAUAUUGGCUGCGAUUUCACGGUCGCUGAUCGCGGUGUUUUAUUUUUUUCAA-UUUUC ..--.............((((((--------------(((((...(((....(((.....)))..(((((......))))).))))))))))))))...........-..... ( -16.30) >consensus ________UU_UUUGCAGGAACUUUGCCAGGAUA___UCCUGUAUAUCCUUUUCAUAUGCUGGCUGCGAUUUCACGCUCGCUGAUCGCAGCGC__UAUUUUUUCCAACAUUUC .................((((......(((((.....)))))....................(((((((((...........)))))))))..........))))........ (-15.62 = -16.40 + 0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:48 2006