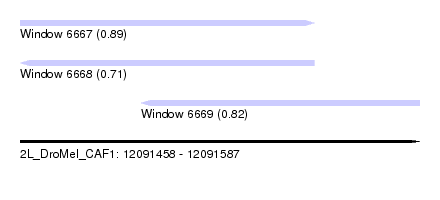
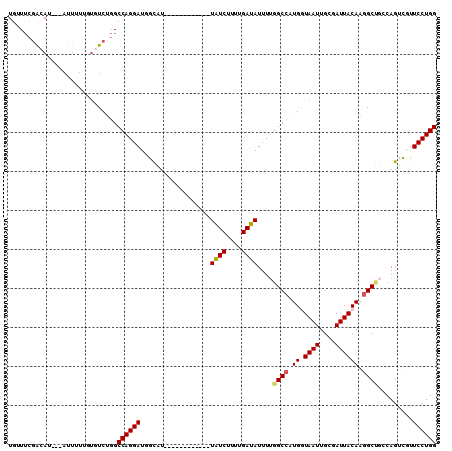
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,091,458 – 12,091,587 |
| Length | 129 |
| Max. P | 0.886842 |

| Location | 12,091,458 – 12,091,553 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 80.59 |
| Mean single sequence MFE | -21.95 |
| Consensus MFE | -15.38 |
| Energy contribution | -15.18 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
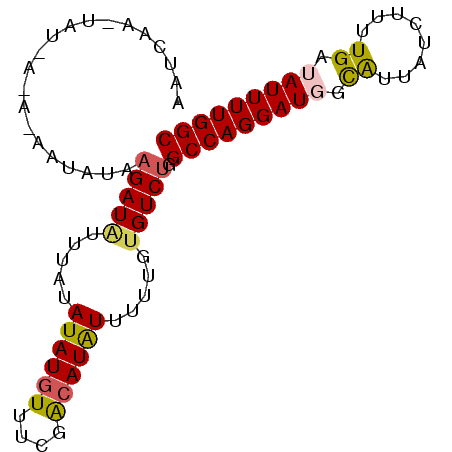
>2L_DroMel_CAF1 12091458 95 + 22407834 CCAGGAAUGACUGGGAGCCUUGUAAUCGCAAUUACCAUGGCCAAAAUAUCAAAAGAUA------------AUGCCAUCCUGGCCAGACACAAAAAU---AUGUCGAAACA ((((((.....((((.(((..(((((....)))))...))))....((((....))))------------...)))))))))...((((.......---.))))...... ( -21.70) >DroSec_CAF1 27871 95 + 1 CCAGGAACGACUGGCAGCCUUGUAAUCGCAAUUACCAUGGCCAAAAUAUCAAAAGAUA------------AUACCAUCCUGGCCAGACACAAAAAU---AUGUCGAAACA ((((((.....(((..(((..(((((....)))))...))).....((((....))))------------...)))))))))...((((.......---.))))...... ( -20.90) >DroSim_CAF1 28163 95 + 1 CCAGGAACGACUGGCAGCCUUGUAAUCGCAAUUACCAUGGCCAAAAUAUCAAAAGAUA------------AUGCCAUCCUGGCCAGACACAAAAAU---AUGUCGAAACA ((((((.....((((((((..(((((....)))))...))).....((((....))))------------.)))))))))))...((((.......---.))))...... ( -26.10) >DroEre_CAF1 28206 93 + 1 CCAGGAACGACUGUCGGCCUUGUAAUCGCAAUUAUCAUGGCCAAAAUAUCAAAAGAUA------------ACGCAAUCCUGGCCAGACA--AAAAC---AUGCCGAAACA ((((((.((......((((.((((((....)))).)).))))....((((....))))------------.))...)))))).......--.....---........... ( -20.20) >DroYak_CAF1 29954 94 + 1 CCAGGAACGUCUGCCAGCCUUGUAAUCGCAAUUACCAUGGCCAAAAUAUCAAAAGAUA------------AUGCAAUCCUGGCCAGACAG-AAAAC---AUCUCGAAACA ........(((((((((..(((((...((..........)).....((((....))))------------.)))))..)))).)))))..-.....---........... ( -19.50) >DroAna_CAF1 29533 110 + 1 CCAGGAAGAGCUGCUGGCUCUGUAAUCGCAAUUAUCACGGCACGAAUAUCAAAAGACAGCGUUUAACAUGACAUGAUCCUGGCGAGAUUGGGAAACGCCAUUUUUUAUUA ((((((...((((((.(((.((((((....)))).)).)))..(.....)...)).))))(((......)))....)))))).((((.(((......))).))))..... ( -23.30) >consensus CCAGGAACGACUGGCAGCCUUGUAAUCGCAAUUACCAUGGCCAAAAUAUCAAAAGAUA____________AUGCCAUCCUGGCCAGACACAAAAAC___AUGUCGAAACA ((((((..........(((..(((((....)))))...))).....((((....))))..................))))))............................ (-15.38 = -15.18 + -0.19)

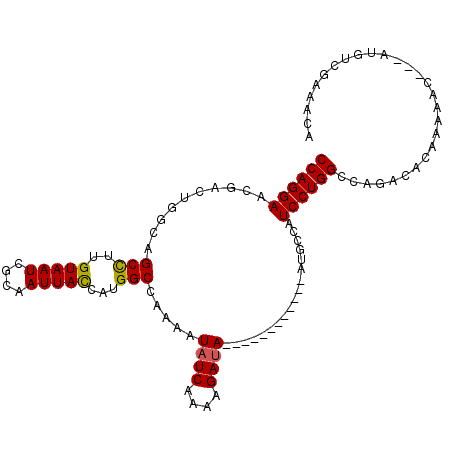
| Location | 12,091,458 – 12,091,553 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 80.59 |
| Mean single sequence MFE | -27.95 |
| Consensus MFE | -16.30 |
| Energy contribution | -16.38 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713302 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12091458 95 - 22407834 UGUUUCGACAU---AUUUUUGUGUCUGGCCAGGAUGGCAU------------UAUCUUUUGAUAUUUUGGCCAUGGUAAUUGCGAUUACAAGGCUCCCAGUCAUUCCUGG ......(((((---(....))))))...((((((((((..------------((((....))))....((((...(((((....)))))..))))....)))).)))))) ( -29.50) >DroSec_CAF1 27871 95 - 1 UGUUUCGACAU---AUUUUUGUGUCUGGCCAGGAUGGUAU------------UAUCUUUUGAUAUUUUGGCCAUGGUAAUUGCGAUUACAAGGCUGCCAGUCGUUCCUGG ......(((((---(....))))))...((((((((((..------------((((....))))....((((...(((((....)))))..)))))))).....)))))) ( -28.10) >DroSim_CAF1 28163 95 - 1 UGUUUCGACAU---AUUUUUGUGUCUGGCCAGGAUGGCAU------------UAUCUUUUGAUAUUUUGGCCAUGGUAAUUGCGAUUACAAGGCUGCCAGUCGUUCCUGG ......(((((---(....))))))...((((((((((..------------((((....))))....((((...(((((....)))))..)))))))).....)))))) ( -30.30) >DroEre_CAF1 28206 93 - 1 UGUUUCGGCAU---GUUUU--UGUCUGGCCAGGAUUGCGU------------UAUCUUUUGAUAUUUUGGCCAUGAUAAUUGCGAUUACAAGGCCGACAGUCGUUCCUGG ....(((((.(---((..(--((((((((((((((..((.------------.......))..)))))))))).)))))..)))........)))))(((......))). ( -28.30) >DroYak_CAF1 29954 94 - 1 UGUUUCGAGAU---GUUUU-CUGUCUGGCCAGGAUUGCAU------------UAUCUUUUGAUAUUUUGGCCAUGGUAAUUGCGAUUACAAGGCUGGCAGACGUUCCUGG ......(..((---(((((-(.(((((((((((((..((.------------.......))..)))))))))...(((((....))))).)))).)).))))))..)... ( -25.30) >DroAna_CAF1 29533 110 - 1 UAAUAAAAAAUGGCGUUUCCCAAUCUCGCCAGGAUCAUGUCAUGUUAAACGCUGUCUUUUGAUAUUCGUGCCGUGAUAAUUGCGAUUACAGAGCCAGCAGCUCUUCCUGG ..........(((......)))......((((((...(((((((....(((.((((....))))..)))..)))))))...........(((((.....))))))))))) ( -26.20) >consensus UGUUUCGACAU___AUUUUUGUGUCUGGCCAGGAUGGCAU____________UAUCUUUUGAUAUUUUGGCCAUGGUAAUUGCGAUUACAAGGCUGCCAGUCGUUCCUGG ............................((((((..................((((....))))....((((.((.((((....)))))).)))).........)))))) (-16.30 = -16.38 + 0.08)

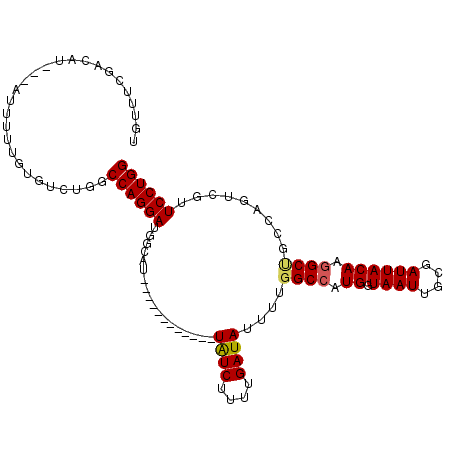
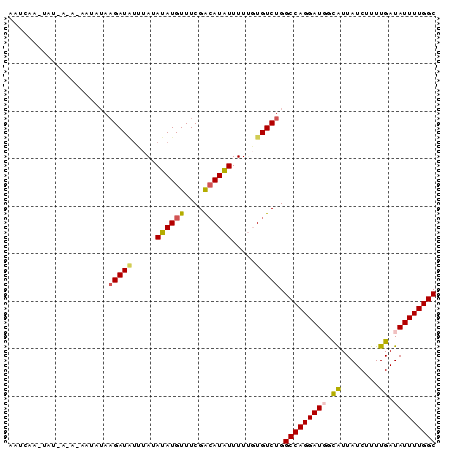
| Location | 12,091,497 – 12,091,587 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 79.64 |
| Mean single sequence MFE | -17.92 |
| Consensus MFE | -14.12 |
| Energy contribution | -14.28 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.816162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12091497 90 - 22407834 AUUCAAGUAUCACAAAAUAUAAGAUAUUUAUAUAUGUUUCGACAUAUUUUUGUGUCUGGCCAGGAUGGCAUUAUCUUUUGAUAUUUUGGC ............((((((((((((((..............((((((....))))))..(((.....)))..))))))...)))))))).. ( -20.10) >DroSec_CAF1 27910 87 - 1 AAUCAAAUAUAAGAUA---UAAGAUAAUUAUAUAUGUUUCGACAUAUUUUUGUGUCUGGCCAGGAUGGUAUUAUCUUUUGAUAUUUUGGC ...........(((((---(((((.......((((((....)))))))))))))))).(((((((((.((........)).))))))))) ( -17.81) >DroSim_CAF1 28202 90 - 1 AAUCAAAUAUAAGAUAUUAUAAGAUAUUUAUAUAUGUUUCGACAUAUUUUUGUGUCUGGCCAGGAUGGCAUUAUCUUUUGAUAUUUUGGC .........((((((((((.((((((..............((((((....))))))..(((.....)))..)))))).)))))))))).. ( -22.90) >DroEre_CAF1 28245 78 - 1 AGCGG----------AGUAUAAGAUGUGUAUAUAUGUUUCGGCAUGUUUU--UGUCUGGCCAGGAUUGCGUUAUCUUUUGAUAUUUUGGC ..(((----------(....(((((((((............)))))))))--..))))((((((((..((........))..)))))))) ( -15.00) >DroYak_CAF1 29993 79 - 1 AACCG----------AAUCAAUGAUCUGCAUAUAUGUUUCGAGAUGUUUU-CUGUCUGGCCAGGAUUGCAUUAUCUUUUGAUAUUUUGGC ..(((----------((((((.(((.((((....((...(.((((.....-..)))).).))....))))..)))..)))))...)))). ( -13.80) >consensus AAUCAA_UAU_A_A_AAUAUAAGAUAUUUAUAUAUGUUUCGACAUAUUUUUGUGUCUGGCCAGGAUGGCAUUAUCUUUUGAUAUUUUGGC .....................(((((.....((((((....)))))).....))))).(((((((((.((........)).))))))))) (-14.12 = -14.28 + 0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:45 2006