| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,083,091 – 12,083,203 |
| Length | 112 |
| Max. P | 0.752932 |

| Location | 12,083,091 – 12,083,203 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -23.41 |
| Consensus MFE | -13.76 |
| Energy contribution | -13.58 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12083091 112 - 22407834 A--AAUAUAAAUAACAUUUUAUUUGUGGUGGAGCAGUACAGACAUUUUCCGUUUGGGAAAACCGCUGACAGACUCGC-UCCUAAACA-AUAAACAUAUGUAUUAGUUCCAAUUCGU .--..(((((((((....))))))))).((((((((((((....(((...((((((((.....((.((.....))))-)))))))).-..)))....)))))).))))))...... ( -25.50) >DroSec_CAF1 19443 112 - 1 A--AAUAUAAAUAACAUUUUAUUUGUGGUGGAGCAGUACAGACAUUUUCCAUUUGGGAAAACCGCUGACAGACUCGC-UCCCGAACA-AUAUACAUAUGUAUUAGUUCCAAUUCGU .--..(((((((((....)))))))))(.((((((((.(((...((((((.....))))))...)))....))).))-))))(((((-(((((....)))))).))))........ ( -27.70) >DroSim_CAF1 19418 111 - 1 A--AACAUAAAUAACAUUUUAUUUGUGGCGGAGCAGUACAGACAUUUUCCAUUUGGGAAAACCGCUGACAGACUCGC-UCCCAAACA-AUAUACAUAUG-AUUAGUUCCAAUUCGU .--..(((((((((....)))))))))(.((((((((.(((...((((((.....))))))...)))....))).))-)))).....-...........-................ ( -24.90) >DroEre_CAF1 19599 98 - 1 A--AGUAAAAAUAACACUU--CUUGU-GUGGAGCAGUACAGACAUUUUCCGCUGGGGAAAACCGCUGACAGACUCGC-UCCCAAACC-AUA-----------UAGUUCCAAUUCGU .--..........((((..--...))-))((((((((.(((...((((((.....))))))...)))....))).))-)))......-...-----------.............. ( -22.10) >DroYak_CAF1 20427 103 - 1 AAAAAUAAAAAUAACAUUUUCUUUGU-GUGGAGCAGUACAGACAUUUUCCGUUUGGGAAAACCGCUGACAGACUCGC-UCCCAAACAAAUA-----------UAGUUCCAAUCCGU .....................(((((-..((((((((.(((...((((((.....))))))...)))....))).))-)))...)))))..-----------.............. ( -22.30) >DroAna_CAF1 21120 94 - 1 G-ACAUGAAAAU-ACAAUUUAUUC---ACGGGGCAGUAUGGACAUUUUCCAAAUG-GAAAACCUCUAAUACACUCGCCUCCCAAA-----A-----------UAGUUCCAAUUCGU .-..(((((...-.....(((((.---..((((((((.((((..((((((....)-)))))..))))....))).)))))....)-----)-----------)))......))))) ( -17.94) >consensus A__AAUAAAAAUAACAUUUUAUUUGU_GUGGAGCAGUACAGACAUUUUCCAUUUGGGAAAACCGCUGACAGACUCGC_UCCCAAACA_AUA___________UAGUUCCAAUUCGU ........................((...((((((((.(((...((((((.....))))))...)))....))).)).)))...)).............................. (-13.76 = -13.58 + -0.18)


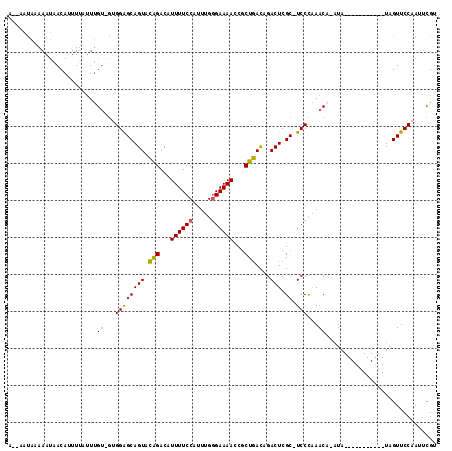
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:38 2006