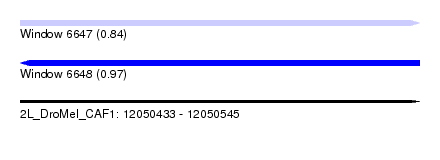
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,050,433 – 12,050,545 |
| Length | 112 |
| Max. P | 0.973190 |

| Location | 12,050,433 – 12,050,545 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 79.32 |
| Mean single sequence MFE | -45.45 |
| Consensus MFE | -28.54 |
| Energy contribution | -29.48 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
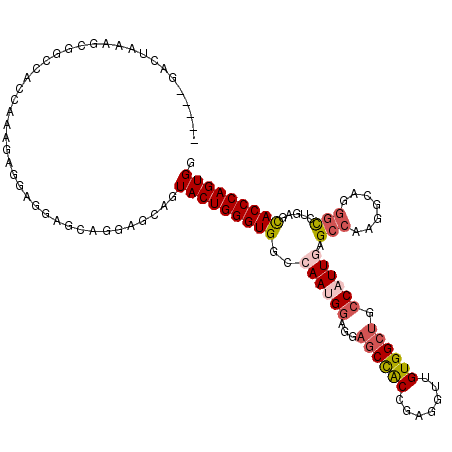
>2L_DroMel_CAF1 12050433 112 + 22407834 -----GACUAAAGCGGCCAGCAAAGAGGAGGAGCAGGAGCAGUACUGGGUGGCCAACGGAGGAGCCACCGAGGUUGUGGCUGCCAUUGAGCCCAGGCAGGGCCUGAGCACCCAGUGG -----.......(((((((.(((.........((....))....((.(((((((.......).)))))).)).))))))))))(((((.((.(((((...))))).))...))))). ( -42.80) >DroSec_CAF1 4596 112 + 1 -----GACUAAAGCGGCCACCAAAGAGGAGGAGCAGGAGCAGUACUGGGUGGCCAAUGGAGGAGCCACCGAGGUUGUGGCUGCCAUUGAGCCCAGGCAGGGCCUGAGCACCCAGUGG -----.......((..((.((.....)).)).))........(((((((((((((((((...((((((.......)))))).)))))).)))(((((...)))))...)))))))). ( -48.40) >DroSim_CAF1 4639 112 + 1 -----GACUAAAGCGGCCACCAAAGAGGAGGAGCAGGAGCAGUACUGGGUGGCCAAUGGAGGAGCCACCGAGGUUGUGGCUGCCAUUGAGCCCAGGCAGGGCCUGAGCACCCAGUGG -----.......((..((.((.....)).)).))........(((((((((((((((((...((((((.......)))))).)))))).)))(((((...)))))...)))))))). ( -48.40) >DroEre_CAF1 4609 109 + 1 -----GACCAAAUCGGACACCAGUGAGGUGGAGCAGAA---GUACUGGGUGGCCAAUGGAGGAGCCACCGAGGUUGUGGCUGCCAUUGAGCCACGAAAGGGUCUGAAUACCCAGUGG -----..((.....))...(((.((.((((...((((.---...((..(((((((((((...((((((.......)))))).)))))).)))))...))..))))..)))))).))) ( -45.40) >DroYak_CAF1 4656 109 + 1 -----UACCAAGACGGCCACCCGUAAGGUGGAGCAGGA---GUACUGGGUGGCCAAUGGAGGAGCCACCGAGGUUGUGGCUGCCAUUCAGCCAAGCCAGGGCCUGACCACCCAGUGG -----..(((..((((....))))..(((((..((((.---...((((.((((.(((((...((((((.......)))))).)))))..))))..))))..)))).)))))...))) ( -47.20) >DroPer_CAF1 4368 108 + 1 UCGCUGGCUAAGUCGACCA---AGCCGCCGUCGUCGCGCCAGUACUGGGUGGCCAACGAGGGUACUGCCGAAAUUGUGGCCGCCAUUCAGCCAAG------CCUCUCCACCCAGUGG ..((((((...(.((((..---.......)))).)..))))))(((((((((.....((((.(...((((......)))).((......))..).------)))).))))))))).. ( -40.50) >consensus _____GACUAAAGCGGCCACCAAAGAGGAGGAGCAGGAGCAGUACUGGGUGGCCAAUGGAGGAGCCACCGAGGUUGUGGCUGCCAUUGAGCCAAGGCAGGGCCUGAGCACCCAGUGG ..........................................(((((((((..((((((...((((((.......)))))).)))))).(((.......))).....))))))))). (-28.54 = -29.48 + 0.95)

| Location | 12,050,433 – 12,050,545 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.32 |
| Mean single sequence MFE | -45.33 |
| Consensus MFE | -30.53 |
| Energy contribution | -31.37 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
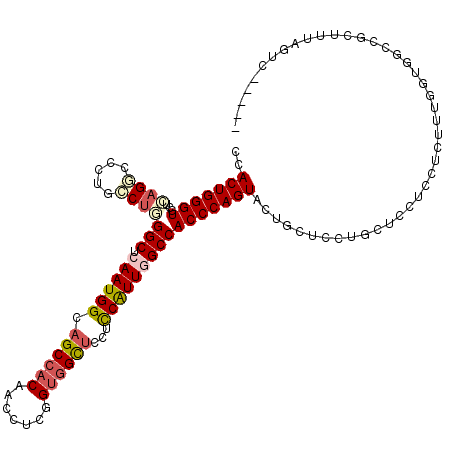
>2L_DroMel_CAF1 12050433 112 - 22407834 CCACUGGGUGCUCAGGCCCUGCCUGGGCUCAAUGGCAGCCACAACCUCGGUGGCUCCUCCGUUGGCCACCCAGUACUGCUCCUGCUCCUCCUCUUUGCUGGCCGCUUUAGUC----- ..(((((((((((((((...))))))))(((((((.((((((.......))))))...)))))))..)))))))...((.((.((...........)).))..)).......----- ( -45.00) >DroSec_CAF1 4596 112 - 1 CCACUGGGUGCUCAGGCCCUGCCUGGGCUCAAUGGCAGCCACAACCUCGGUGGCUCCUCCAUUGGCCACCCAGUACUGCUCCUGCUCCUCCUCUUUGGUGGCCGCUUUAGUC----- ..(((((((((((((((...))))))))(((((((.((((((.......))))))...)))))))..)))))))...(((...((.((.((.....)).))..))...))).----- ( -48.20) >DroSim_CAF1 4639 112 - 1 CCACUGGGUGCUCAGGCCCUGCCUGGGCUCAAUGGCAGCCACAACCUCGGUGGCUCCUCCAUUGGCCACCCAGUACUGCUCCUGCUCCUCCUCUUUGGUGGCCGCUUUAGUC----- ..(((((((((((((((...))))))))(((((((.((((((.......))))))...)))))))..)))))))...(((...((.((.((.....)).))..))...))).----- ( -48.20) >DroEre_CAF1 4609 109 - 1 CCACUGGGUAUUCAGACCCUUUCGUGGCUCAAUGGCAGCCACAACCUCGGUGGCUCCUCCAUUGGCCACCCAGUAC---UUCUGCUCCACCUCACUGGUGUCCGAUUUGGUC----- (((((((((.....((.....))..(((.((((((.((((((.......))))))...))))))))))))))))..---........((((.....))))........))..----- ( -39.00) >DroYak_CAF1 4656 109 - 1 CCACUGGGUGGUCAGGCCCUGGCUUGGCUGAAUGGCAGCCACAACCUCGGUGGCUCCUCCAUUGGCCACCCAGUAC---UCCUGCUCCACCUUACGGGUGGCCGUCUUGGUA----- ..(((((((((((.((((.......)))).(((((.((((((.......))))))...))))))))))))))))..---.((.((.(((((.....)))))..))...))..----- ( -48.30) >DroPer_CAF1 4368 108 - 1 CCACUGGGUGGAGAGG------CUUGGCUGAAUGGCGGCCACAAUUUCGGCAGUACCCUCGUUGGCCACCCAGUACUGGCGCGACGACGGCGGCU---UGGUCGACUUAGCCAGCGA ..(((((((((.((((------....(((((((((...)))....)))))).....)))).....))))))))).(((((.((((.(.(....).---).)))).....)))))... ( -43.30) >consensus CCACUGGGUGCUCAGGCCCUGCCUGGGCUCAAUGGCAGCCACAACCUCGGUGGCUCCUCCAUUGGCCACCCAGUACUGCUCCUGCUCCUCCUCUUUGGUGGCCGCUUUAGUC_____ ..(((((((...((((.....))))(((.((((((.((((((.......))))))...))))))))))))))))........................................... (-30.53 = -31.37 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:24 2006