
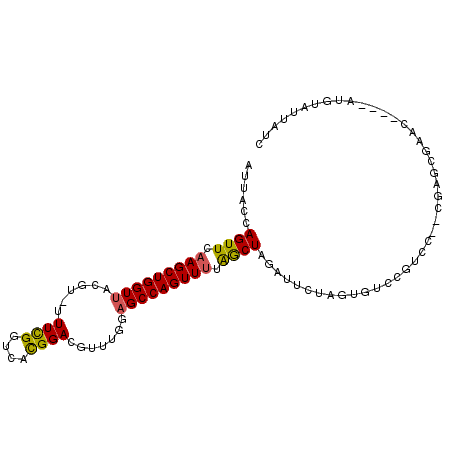
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,024,370 – 12,024,471 |
| Length | 101 |
| Max. P | 0.962261 |

| Location | 12,024,370 – 12,024,471 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 79.40 |
| Mean single sequence MFE | -32.98 |
| Consensus MFE | -15.15 |
| Energy contribution | -14.70 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
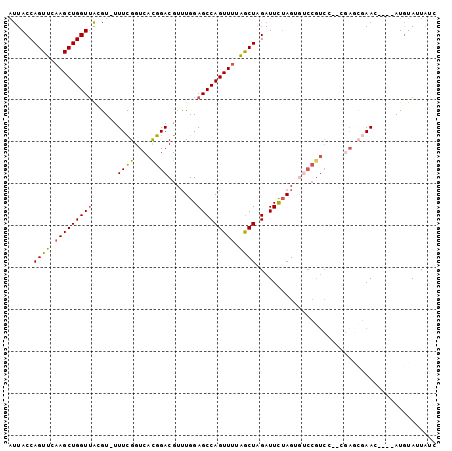
>2L_DroMel_CAF1 12024370 101 + 22407834 AUUACCAGUUCAAGCUGGUUAAGU-UUUCGGUCACGGACGUUUGGAGCCAGUUUUGGCUAGAUUCUAGUAUCCGUCCGUCGAGCGAACAUGUAUGUAUUAUC ...((((((....)))))).....-.((((.(((((((((.....((((......)))).(((......)))))))))).)).))))............... ( -31.50) >DroSec_CAF1 15759 101 + 1 AUUACCAGUUCAAGCUGGUUACGU-UUUCGGUCACGGACGUUUGGAGCCAGUUUUGACUAGAUUCUAGUGUCCGUCCGUCGAGCGAAAACGUAUGUAUUAUC ...((((((....))))))(((((-(((((.(((((((((.(((((((.(((....))).).))))))....))))))).)).))))))))))......... ( -39.70) >DroSim_CAF1 15903 101 + 1 AUUACCAGUUCAAGCUGGUUACGU-UUUCGGUCACGGACGUUUGGAGCCAGUUUUGGCUAGAUUCUAGUGUCCGUCCGUCGAGCGAAAACGUAUGUAUUAUC ...((((((....))))))(((((-(((((.(((((((((.....((((......)))).(((......)))))))))).)).))))))))))......... ( -41.80) >DroEre_CAF1 16174 89 + 1 ACUACCAGUUCAAGCUGGUUGGGU-UUUCGGUCAUGGACGUUUGGAGCCAGUUUAAGCUGGAUUCUAGAGUCCGUCG--------AAC----AUGUAUUACC .(.((((((....)))))).)(((-...((.((((((((.((((((((((((....))))).))))))))))))).)--------)..----.))....))) ( -32.30) >DroYak_CAF1 16560 95 + 1 AUUACCAGUUCAAGCUGGUUACGU-UUUCGGUCAUGGACGUUUGGAGCCAGUUUAAGCUGGAUUCUAGAGCCCGUCG--AGAGCGAAC----AUGUAUUAUA ...((((((....))))))(((((-.((((.((.(.((((((((((((((((....))))).)))))))...)))).--))).)))).----)))))..... ( -35.30) >DroAna_CAF1 18254 83 + 1 UUUACCAGUUCAAGCUGGUAAAAUUUUUUGGUCAUGGACGU-UGGAGCCAGUUAUAGCUAGAUUU-AG-------------ACAUAAC----AUAUAUUUAC (((((((((....)))))))))............(((.(..-....))))(((((..(((....)-))-------------..)))))----.......... ( -17.30) >consensus AUUACCAGUUCAAGCUGGUUACGU_UUUCGGUCACGGACGUUUGGAGCCAGUUUUAGCUAGAUUCUAGUGUCCGUCC__CGAGCGAAC____AUGUAUUAUC ......((((.(((((((((......((((....)))).......))))))))).))))........................................... (-15.15 = -14.70 + -0.44)

| Location | 12,024,370 – 12,024,471 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 79.40 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -12.11 |
| Energy contribution | -12.34 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.876865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
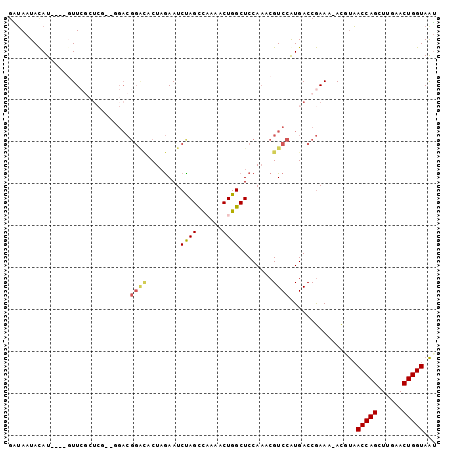
>2L_DroMel_CAF1 12024370 101 - 22407834 GAUAAUACAUACAUGUUCGCUCGACGGACGGAUACUAGAAUCUAGCCAAAACUGGCUCCAAACGUCCGUGACCGAAA-ACUUAACCAGCUUGAACUGGUAAU ...............((((.((.((((((((((......))).(((((....))))).....))))))))).)))).-.....(((((......)))))... ( -29.20) >DroSec_CAF1 15759 101 - 1 GAUAAUACAUACGUUUUCGCUCGACGGACGGACACUAGAAUCUAGUCAAAACUGGCUCCAAACGUCCGUGACCGAAA-ACGUAACCAGCUUGAACUGGUAAU .........((((((((((.((.(((((((...(((((...)))))......((....))..))))))))).)))))-)))))(((((......)))))... ( -35.50) >DroSim_CAF1 15903 101 - 1 GAUAAUACAUACGUUUUCGCUCGACGGACGGACACUAGAAUCUAGCCAAAACUGGCUCCAAACGUCCGUGACCGAAA-ACGUAACCAGCUUGAACUGGUAAU .........((((((((((.((.(((((((((........)).(((((....))))).....))))))))).)))))-)))))(((((......)))))... ( -37.50) >DroEre_CAF1 16174 89 - 1 GGUAAUACAU----GUU--------CGACGGACUCUAGAAUCCAGCUUAAACUGGCUCCAAACGUCCAUGACCGAAA-ACCCAACCAGCUUGAACUGGUAGU (((....(((----(..--------((..(((........)))((((......)))).....))..)))))))....-.....(((((......)))))... ( -19.50) >DroYak_CAF1 16560 95 - 1 UAUAAUACAU----GUUCGCUCU--CGACGGGCUCUAGAAUCCAGCUUAAACUGGCUCCAAACGUCCAUGACCGAAA-ACGUAACCAGCUUGAACUGGUAAU .....(((..----.((((.((.--.((((.......((..((((......)))).))....))))...)).)))).-..)))(((((......)))))... ( -20.40) >DroAna_CAF1 18254 83 - 1 GUAAAUAUAU----GUUAUGU-------------CU-AAAUCUAGCUAUAACUGGCUCCA-ACGUCCAUGACCAAAAAAUUUUACCAGCUUGAACUGGUAAA ..........----((((((.-------------..-......(((((....)))))...-.....))))))........((((((((......)))))))) ( -15.73) >consensus GAUAAUACAU____GUUCGCUCG__GGACGGACACUAGAAUCUAGCCAAAACUGGCUCCAAACGUCCAUGACCGAAA_ACGUAACCAGCUUGAACUGGUAAU .............................((((........((((......))))........))))................(((((......)))))... (-12.11 = -12.34 + 0.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:18 2006