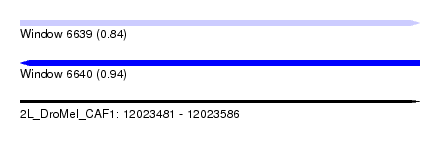
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,023,481 – 12,023,586 |
| Length | 105 |
| Max. P | 0.941598 |

| Location | 12,023,481 – 12,023,586 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 78.90 |
| Mean single sequence MFE | -25.52 |
| Consensus MFE | -11.38 |
| Energy contribution | -11.46 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12023481 105 + 22407834 UUAUUCUAUAUCCUAAGCUGACUACUCUUCUAUGGAAUUUGAGCUAGAAUUUAGUCAGCGUGAUUAGCUUCAGAUACAGGAUA-UCUUAUCGCGACAGAUAUAAUU .((((((.((((....((((((((...(((((............)))))..)))))))).(((......))))))).))))))-...((((......))))..... ( -23.80) >DroSec_CAF1 14841 95 + 1 AUAUUCUAUAUCCCAAGCUGACUACUCUGCUAUGGAAUUUUAGC-----------CAGCGUGAUUAGCUGCAGAUACAGUAUCUUCUUAUCGCGACAGAUAUAAUU ......((((((...((((((.(((.((((((........))).-----------))).))).))))))((.((((.((......))))))))....))))))... ( -22.40) >DroSim_CAF1 14956 106 + 1 UAAUUCUAUAUCCCAAGCUGACUGCUCUGCUAUGGAAUUUUAGCUAGAAUUUAGUCAGCUUGAUUAGCUGCAGAUACAGUAUCUUCUUAUCGCGACAGAUAUAAUU ......((((((.(((((((((((.(((((((........)))).)))...)))))))))))....(.(((.((((.((......))))))))).).))))))... ( -31.10) >DroEre_CAF1 15303 92 + 1 UCAUUCUAUAUCAUGAGCU-------------UGGCAUUUUAAGCACAGUUUAGGCAGCGUGAUCAGCUGCAGAUAAAGGAUA-UCUUAUCGCGACGGAUAUAACU ..(((((.((((.((((((-------------..((.......))..)))))).(((((.......))))).)))).))))).-...((((......))))..... ( -22.30) >DroYak_CAF1 15681 105 + 1 CUAUUCUAUAUUCCAAGCUGACUACUCUGCUGUGGAAUUUUAGCCGCAAUUUAGGCGGCGUGAUUAGCUGCAGAUAAAGGAUA-UCUUAUCGCGACAGAUACAAUU .((((((.((((...((((((((((......)))).......(((((.......)))))....))))))...)))).))))))-...((((......))))..... ( -28.00) >consensus UUAUUCUAUAUCCCAAGCUGACUACUCUGCUAUGGAAUUUUAGCCACAAUUUAGGCAGCGUGAUUAGCUGCAGAUACAGGAUA_UCUUAUCGCGACAGAUAUAAUU ......((((((....((.....................................((((.......))))..((((.((......))))))))....))))))... (-11.38 = -11.46 + 0.08)

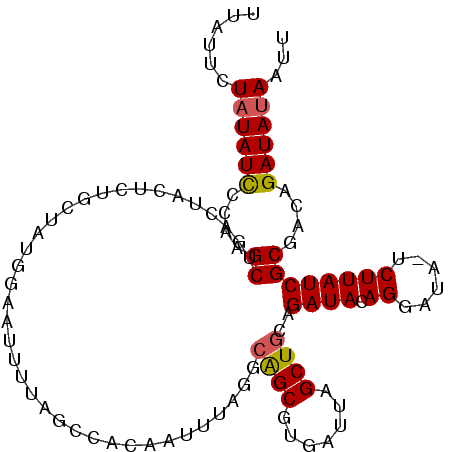
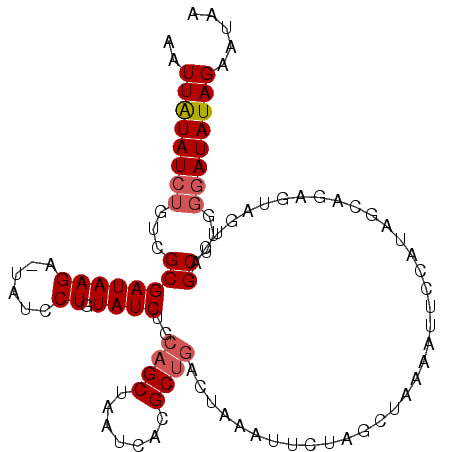
| Location | 12,023,481 – 12,023,586 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 78.90 |
| Mean single sequence MFE | -25.70 |
| Consensus MFE | -10.70 |
| Energy contribution | -11.34 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.42 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12023481 105 - 22407834 AAUUAUAUCUGUCGCGAUAAGA-UAUCCUGUAUCUGAAGCUAAUCACGCUGACUAAAUUCUAGCUCAAAUUCCAUAGAAGAGUAGUCAGCUUAGGAUAUAGAAUAA ..((((((((...((....(((-(((...))))))...)).......((((((((..(((((............)))))...))))))))...))))))))..... ( -28.90) >DroSec_CAF1 14841 95 - 1 AAUUAUAUCUGUCGCGAUAAGAAGAUACUGUAUCUGCAGCUAAUCACGCUG-----------GCUAAAAUUCCAUAGCAGAGUAGUCAGCUUGGGAUAUAGAAUAU .....(((((.((.......)))))))((((((((..((((.((.((.(((-----------.(((........)))))).)).)).))))..))))))))..... ( -23.10) >DroSim_CAF1 14956 106 - 1 AAUUAUAUCUGUCGCGAUAAGAAGAUACUGUAUCUGCAGCUAAUCAAGCUGACUAAAUUCUAGCUAAAAUUCCAUAGCAGAGCAGUCAGCUUGGGAUAUAGAAUUA ..((((((((((.((((((((......)).)))).)).))....((((((((((...((((.((((........)))))))).))))))))))))))))))..... ( -32.40) >DroEre_CAF1 15303 92 - 1 AGUUAUAUCCGUCGCGAUAAGA-UAUCCUUUAUCUGCAGCUGAUCACGCUGCCUAAACUGUGCUUAAAAUGCCA-------------AGCUCAUGAUAUAGAAUGA ..(((((((....((((((((.-.....)))))).(((((.......)))))......((.((.......))))-------------.))....)))))))..... ( -18.00) >DroYak_CAF1 15681 105 - 1 AAUUGUAUCUGUCGCGAUAAGA-UAUCCUUUAUCUGCAGCUAAUCACGCCGCCUAAAUUGCGGCUAAAAUUCCACAGCAGAGUAGUCAGCUUGGAAUAUAGAAUAG .......(((((.((....(((-((.....)))))...)).......(((((.......)))))....((((((.(((.((....)).)))))))))))))).... ( -26.10) >consensus AAUUAUAUCUGUCGCGAUAAGA_UAUCCUGUAUCUGCAGCUAAUCACGCUGACUAAAUUCUAGCUAAAAUUCCAUAGCAGAGUAGUCAGCUUGGGAUAUAGAAUAA ..((((((((...((((((((......)).))))..((((.......)))).....................................))...))))))))..... (-10.70 = -11.34 + 0.64)

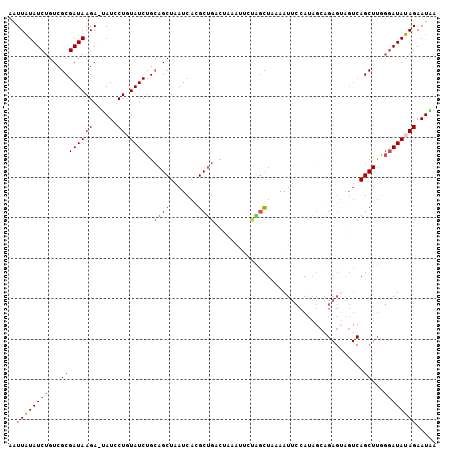
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:16 2006