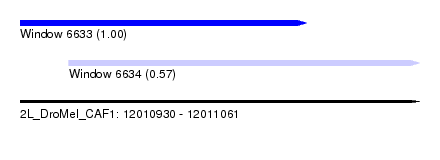
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,010,930 – 12,011,061 |
| Length | 131 |
| Max. P | 0.995505 |

| Location | 12,010,930 – 12,011,024 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 95.22 |
| Mean single sequence MFE | -22.58 |
| Consensus MFE | -21.80 |
| Energy contribution | -21.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.995505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
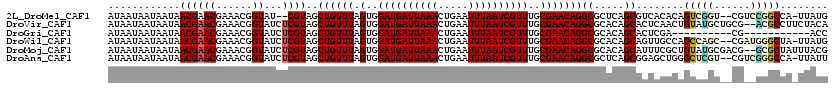
>2L_DroMel_CAF1 12010930 94 + 22407834 C----GACUUGUUGCAUGUAAUAAUAAUAAUAACGAACGAAACGGUAU--CGUAGCUGUUUAGUGGAUGAUUAAACUGAAUUUAAUCGUUUGCGAACAGG .----...((((((....))))))........((((((......)).)--)))..((((((.(..((((((((((.....))))))))))..))))))). ( -22.00) >DroVir_CAF1 2555 100 + 1 AAGACGACUUGUUGCAUGUAAUAAUAAUAAUAACGAACGAAACGGUAUCUCGUAGCUGUUUAGUGGAUGAUUAAACUGAAUUUAAUCGUUUGCGAACAGG .......((((((.(........................(((((((........))))))).(..((((((((((.....))))))))))..)))))))) ( -21.80) >DroGri_CAF1 2718 100 + 1 UGGCCGACUUGUUGCAUGUAAUAAUAAUAAUAACGAACGAAACGGUAUCUCGUAGCUGUUUAGUGGAUGAUUAAACUGAAUUUAAUCGUUUGCGAACAGG ..((((..(((((...(((.((.......)).))))))))..)))).........((((((.(..((((((((((.....))))))))))..))))))). ( -23.70) >DroSec_CAF1 2423 97 + 1 CGG-GGACUUGUUGCAUGUAAUAAUAAUAAUAACGAACGAAACGGUAU--CGUAGCUGUUUAGUGGAUGAUUAAACUGAAUUUAAUCGUUUGCGAACAGG .(.-...)((((((....))))))........((((((......)).)--)))..((((((.(..((((((((((.....))))))))))..))))))). ( -23.30) >DroSim_CAF1 2424 95 + 1 CG---GACUUGUUGCAUGUAAUAAUAAUAAUAACGAACGAAACGGUAU--CGUAGCUGUUUAGUGGAUGAUUAAACUGAAUUUAAUCGUUUGCGAACAGG ..---...((((((....))))))........((((((......)).)--)))..((((((.(..((((((((((.....))))))))))..))))))). ( -22.00) >DroWil_CAF1 3032 97 + 1 --G-AGACUUGUUGCAUGUAAUAAUAAUAAUAACGAACGAAACGGUAUCUCGUAGCUGUUUAGUGGAUGAUUAAACUGAAUUUAAUCGUUUGCGAACAGG --(-(((.((((((....)))))).........((.......))...))))....((((((.(..((((((((((.....))))))))))..))))))). ( -22.70) >consensus CGG__GACUUGUUGCAUGUAAUAAUAAUAAUAACGAACGAAACGGUAU__CGUAGCUGUUUAGUGGAUGAUUAAACUGAAUUUAAUCGUUUGCGAACAGG .......((((((.(........................(((((((........))))))).(..((((((((((.....))))))))))..)))))))) (-21.80 = -21.80 + 0.00)


| Location | 12,010,946 – 12,011,061 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.11 |
| Mean single sequence MFE | -31.55 |
| Consensus MFE | -21.08 |
| Energy contribution | -21.73 |
| Covariance contribution | 0.66 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
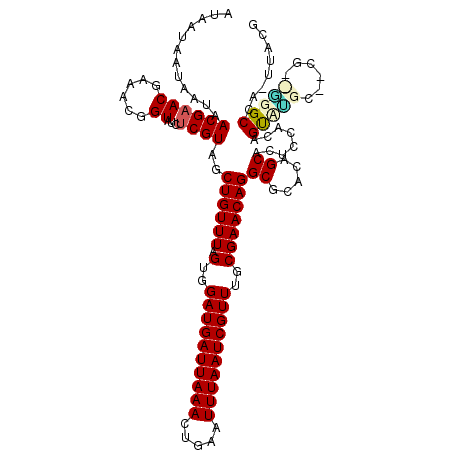
>2L_DroMel_CAF1 12010946 115 + 22407834 AUAAUAAUAAUAACGAACGAAACGGUAU--CGUAGCUGUUUAGUGGAUGAUUAAACUGAAUUUAAUCGUUUGCGAACAGGCGCUCAGCGUCACACAGUCGGU--CGUCCGGCCA-UUAUG ......(((((.((((((......)).)--))).(((((...(..((((((((((.....))))))))))..).....(((((...)))))..))))).(((--(....)))))-)))). ( -31.10) >DroVir_CAF1 2575 118 + 1 AUAAUAAUAAUAACGAACGAAACGGUAUCUCGUAGCUGUUUAGUGGAUGAUUAAACUGAAUUUAAUCGUUUGCGAACAGGCGCACAGCACUCAACUGUAUGCUGCG--ACGCCUUCUACA ...................(((((((........))))))).(..((((((((((.....))))))))))..)((..((((((.(((((..(....)..))))).)--.))))))).... ( -33.70) >DroGri_CAF1 2738 99 + 1 AUAAUAAUAAUAACGAACGAAACGGUAUCUCGUAGCUGUUUAGUGGAUGAUUAAACUGAAUUUAAUCGUUUGCGAACAGGCGCACAGCACUCGA----------CG-----------ACC .......................(((...(((.((((((((.(..((((((((((.....))))))))))..)))))))((.....)).)))))----------..-----------))) ( -24.40) >DroWil_CAF1 3049 117 + 1 AUAAUAAUAAUAACGAACGAAACGGUAUCUCGUAGCUGUUUAGUGGAUGAUUAAACUGAAUUUAAUCGUUUGCGAACAGGCGCACAGCAGUUGCCAGCCAGC--CGAUGGGCUA-UUAUG ......((((((...........((((.((.....((((((.(..((((((((((.....))))))))))..)))))))((.....)))).)))).....((--(....)))))-)))). ( -30.70) >DroMoj_CAF1 2763 118 + 1 AUAAUAAUAAUAACGAACGAAACGGUAUCUCGUAGCUGUUUAGUGGAUGAUUAAACUGAAUUUAAUCGUUUGCGAACAGGCGCACAGCAUUUCGCUGUAUGCGACG--GCGCUAUUUACG .................(((((..((..((((((.((((((.(..((((((((((.....))))))))))..)))))))....(((((.....))))).))))).)--..))..))).)) ( -33.80) >DroAna_CAF1 2532 117 + 1 AUAAUAAUAAUAACGAACGAAACGGUAUCUCGUAGCUGUUUAGUGGAUGAUUAAACUGAAUUUAAUCGUUUGCGAACAGGCGCUCAGCGGAGCUGGGCUCGU--CGUCGGGCCA-UUAUU ..((((((.....(((.(((.((((....))))..((((((.(..((((((((((.....))))))))))..)))))))(.(((((((...))))))).).)--)))))....)-))))) ( -35.60) >consensus AUAAUAAUAAUAACGAACGAAACGGUAUCUCGUAGCUGUUUAGUGGAUGAUUAAACUGAAUUUAAUCGUUUGCGAACAGGCGCACAGCACUCCACAGUAUGC__CG__GGGCCA_UUACG ............((((((......))...))))..((((((.(..((((((((((.....))))))))))..)))))))((.....))........(((((......)))))........ (-21.08 = -21.73 + 0.66)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:10 2006