
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
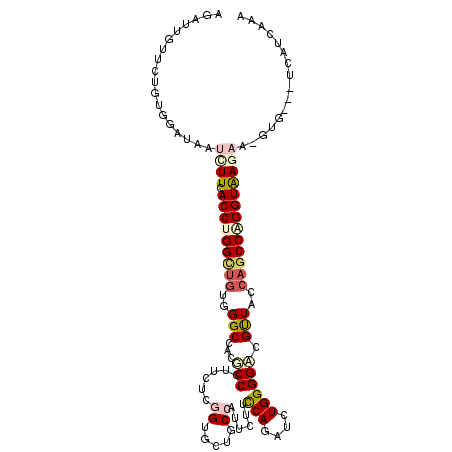
| Location | 12,008,999 – 12,009,110 |
| Length | 111 |
| Max. P | 0.930935 |

| Location | 12,008,999 – 12,009,110 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 71.75 |
| Mean single sequence MFE | -34.75 |
| Consensus MFE | -18.42 |
| Energy contribution | -19.40 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
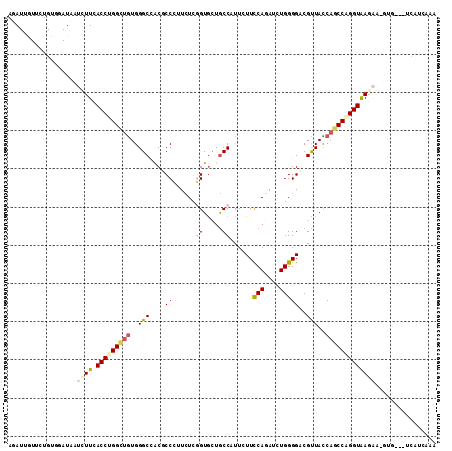
>2L_DroMel_CAF1 12008999 111 + 22407834 AGAUUGUACUGCUGAUACUCUUCACCUGGUUGUGGGCCACUCCGUUCUCCGUGCUGCCAUUGUUCCAGAUCUGGGGACGUUACCAGCCGGGUGAGAA-GUGAAUUCAUGAAG ...............((((..(((((((((((.(((((((..........)))..)))...(((((.......)))))....))))))))))))..)-)))........... ( -34.00) >DroVir_CAF1 583 103 + 1 AGAUCAUUAUGUGCAUCAUAUUCACCUGGCUGUGGGCAACGCCGUUUUCGGUGCUGCCCCUCUUCCAGAUCUGGGGUCGCUACCAGCCAGGUAAC---------UCUUCCCA .((....((((.....)))).))(((((((((.(((((.(((((....))))).)))))...(((((....))))).......)))))))))...---------........ ( -39.30) >DroEre_CAF1 556 111 + 1 AAAUUGUGCUGCUAAUACUCUUCACCUGGCUGUGGGCCGCUCCGUUCUCCGUGCUGCCAUUGUUCCAGAUCUGGGGACGUUACCAGCCGGGUAAGAA-GUGGAUACAUCAGG ....(((((..((.....((((.(((((((((.((((.((..((.....)).)).)))...(((((.......)))))....)))))))))))))))-)..).))))..... ( -38.30) >DroWil_CAF1 556 108 + 1 AAAUUCUUUUGUGUAUUAUAUUUACCUGGCUAUGGGCCACACCCUUCUCGGUAUUGCCAUUCUUUCAGAUAUGGGGGCGUUAUCAGCCUGGUAAGCAAUUG----CAUAGAA .......((((((((.....((((((.((((..(((.....))).....((((.((((.(((..........))))))).)))))))).))))))....))----)))))). ( -31.10) >DroMoj_CAF1 579 107 + 1 AGAUAAUUUUGUGUAUUAUUUUCACCUGGAUGUGGGCAACGCCCUUCUCGGUGCUGCCGCUCUUUCAGAUCUGGGGUCGCUAUCUGCCAGGUGAGAU-GU----UCAUGAAA ......(((..((.((...((((((((((.....((((.((((......)))).))))((..(..((....))..)..))......)))))))))).-))----.))..))) ( -36.30) >DroAna_CAF1 557 104 + 1 AGAUAGUCCUGUUGAUCAUCUUCACCUGGCUGUGGGCCACCCCCUUUUCGGUGCUACCUUUAUUCCAAAUUUGGGGACGUUAUCAGCCCGGUAAGAA-GCGGGUA------- .......((((((.....((((.(((.(((((.((..((((........))))...))....(((((....))))).......))))).))))))))-)))))..------- ( -29.50) >consensus AGAUUGUUCUGUGGAUAAUCUUCACCUGGCUGUGGGCCACGCCCUUCUCGGUGCUGCCAUUCUUCCAGAUCUGGGGACGUUACCAGCCAGGUAAGAA_GUG___UCAUCAAA ..................((((.(((((((((..(((...(((......((.....))......(((....)))))).)))..)))))))))))))................ (-18.42 = -19.40 + 0.98)

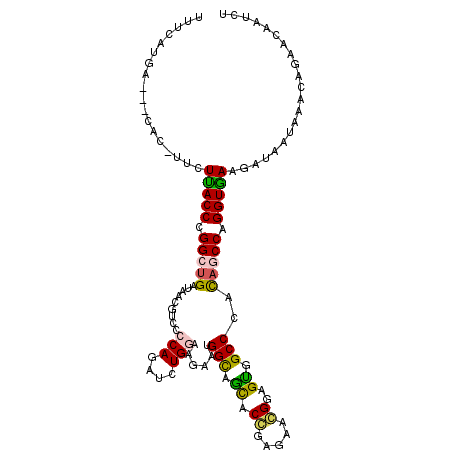
| Location | 12,008,999 – 12,009,110 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 71.75 |
| Mean single sequence MFE | -31.62 |
| Consensus MFE | -20.87 |
| Energy contribution | -20.48 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
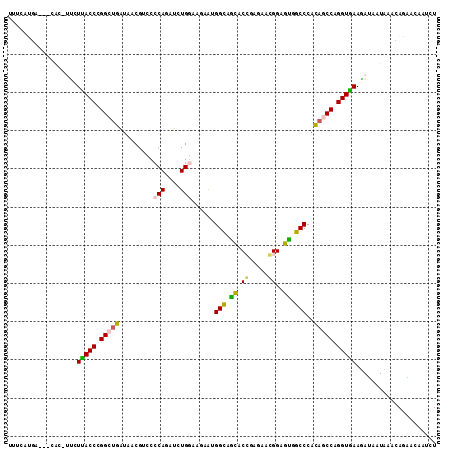
>2L_DroMel_CAF1 12008999 111 - 22407834 CUUCAUGAAUUCAC-UUCUCACCCGGCUGGUAACGUCCCCAGAUCUGGAACAAUGGCAGCACGGAGAACGGAGUGGCCCACAACCAGGUGAAGAGUAUCAGCAGUACAAUCU ((((.......(((-(((...((((((((.((..((..(((....))).))..)).)))).))).)...))))))(((........))))))).((((.....))))..... ( -28.00) >DroVir_CAF1 583 103 - 1 UGGGAAGA---------GUUACCUGGCUGGUAGCGACCCCAGAUCUGGAAGAGGGGCAGCACCGAAAACGGCGUUGCCCACAGCCAGGUGAAUAUGAUGCACAUAAUGAUCU ..(((...---------.(((((((((((.........(((....))).....(((((((.(((....))).))))))).))))))))))).((((.....))))....))) ( -42.50) >DroEre_CAF1 556 111 - 1 CCUGAUGUAUCCAC-UUCUUACCCGGCUGGUAACGUCCCCAGAUCUGGAACAAUGGCAGCACGGAGAACGGAGCGGCCCACAGCCAGGUGAAGAGUAUUAGCAGCACAAUUU ..((.(((....((-(..(((((.(((((.....((..(((....))).))...(((.((.((.....))..)).)))..))))).)))))..)))....))).))...... ( -33.60) >DroWil_CAF1 556 108 - 1 UUCUAUG----CAAUUGCUUACCAGGCUGAUAACGCCCCCAUAUCUGAAAGAAUGGCAAUACCGAGAAGGGUGUGGCCCAUAGCCAGGUAAAUAUAAUACACAAAAGAAUUU ((((..(----(....))(((((.(((((.....(((.((((.((.....))))))..(((((......))))))))...))))).)))))..............))))... ( -25.30) >DroMoj_CAF1 579 107 - 1 UUUCAUGA----AC-AUCUCACCUGGCAGAUAGCGACCCCAGAUCUGAAAGAGCGGCAGCACCGAGAAGGGCGUUGCCCACAUCCAGGUGAAAAUAAUACACAAAAUUAUCU ........----..-...(((((((((((((...........))))).....(.((((((.((......)).)))))))....))))))))..................... ( -30.00) >DroAna_CAF1 557 104 - 1 -------UACCCGC-UUCUUACCGGGCUGAUAACGUCCCCAAAUUUGGAAUAAAGGUAGCACCGAAAAGGGGGUGGCCCACAGCCAGGUGAAGAUGAUCAACAGGACUAUCU -------.......-...(((((.(((((....(..((((...(((((.............)))))..))))..).....))))).)))))(((((.((.....)).))))) ( -30.32) >consensus UUUCAUGA___CAC_UUCUUACCCGGCUGAUAACGUCCCCAGAUCUGGAAGAAUGGCAGCACCGAGAACGGAGUGGCCCACAGCCAGGUGAAGAUAAUAAACAGAACAAUCU ..................(((((.(((((.........(((....)))......(((.((.((.....))..)).)))..))))).)))))..................... (-20.87 = -20.48 + -0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:07 2006