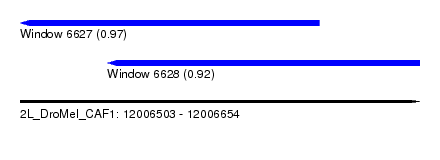
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,006,503 – 12,006,654 |
| Length | 151 |
| Max. P | 0.973895 |

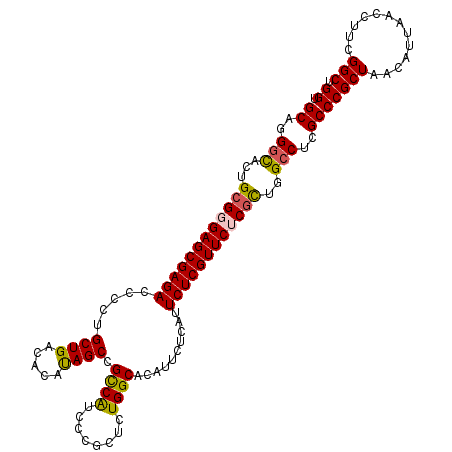
| Location | 12,006,503 – 12,006,616 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 94.15 |
| Mean single sequence MFE | -39.23 |
| Consensus MFE | -35.14 |
| Energy contribution | -35.06 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12006503 113 - 22407834 GCAGGGUACUGCGGGAGCGAGACCCCUGCUGACACAUAGCCGUCGUCCCGCUCUGGCACAUUCUCAUUCUCGUUCUCGUUGGCCUCGCCCGCUAACAUUAACCUUCGGCUGGU ((.((((..(((.((((((.(((..(.((((.....)))).)..))).)))))).)))...................((((((.......))))))....))))...)).... ( -35.70) >DroSec_CAF1 7226 113 - 1 GCAGGUCACUGCGGGAGCGAGACCCCUGCUGACACAUAGCCGCCAUCCCGCUCUGGCACAUUCUCAUUCUCGUUCUCACUGGCCUCGCCCGCUAACAUUAACCUUCGGCUGGU ((((....))))((((((((((.....((((.....)))).((((........))))..........))))))))))((..(((......................)))..)) ( -36.05) >DroSim_CAF1 6439 113 - 1 GCAGGGCACUGCGGGAGCGAGACCCCUGCUGACACAUAGCCGCCAUCCCGCUCUGGCACAUUCUCAUUCUCGUUCUCGCUGGCCUCGCCCGCUAACAUUAACCUUCGGCUGGU ((..(((...((((((((((((.....((((.....)))).((((........))))..........))))))))))))..)))..))(((((.............))).)). ( -40.12) >DroEre_CAF1 6482 109 - 1 GCAGGGCAUUGCGUGAGCGAGACCCCUGCUGACA--CAGCCGCCAUCCCGCUCUGGC--AUUCUCAUUCUCGUUCUCGCUGGCCUCGCCCGCUAGCAUUAACCUUCGGCUGGU ((((((..((((....))))...)))))).....--(((((((((........))))--..................((((((.......))))))..........))))).. ( -43.80) >DroYak_CAF1 6580 111 - 1 GCAGGGCAUUGCGUGAGCGAGACCCCUGCUGACACACAGCCGCCAUCCCGCUCUGGC--AUUCUCAUUCUCGUUCUCGCUGGCCUCGCCCGCUAACAUUAACCUUCGGCUGGU ((((((..((((....))))...)))))).......(((((((((........))))--............(((...((.(((...))).)).)))..........))))).. ( -40.50) >consensus GCAGGGCACUGCGGGAGCGAGACCCCUGCUGACACAUAGCCGCCAUCCCGCUCUGGCACAUUCUCAUUCUCGUUCUCGCUGGCCUCGCCCGCUAACAUUAACCUUCGGCUGGU ((..(((...((((((((((((.....((((.....)))).((((........))))..........))))))))))))..)))..))(((((.............))).)). (-35.14 = -35.06 + -0.08)


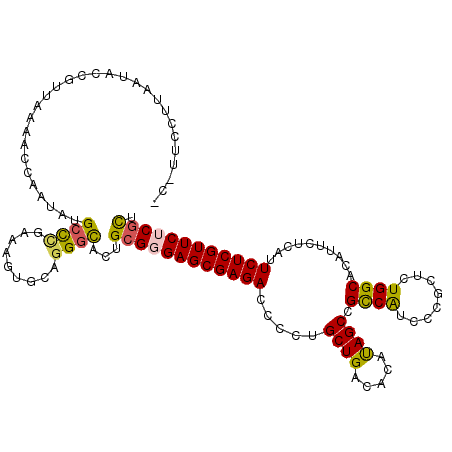
| Location | 12,006,536 – 12,006,654 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 86.61 |
| Mean single sequence MFE | -33.10 |
| Consensus MFE | -27.46 |
| Energy contribution | -27.26 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12006536 118 - 22407834 CC-UUCCUUAAUACCGUUUAAGCCAAUAUGCUCGAAAGUGCAGGGUACUGCGGGAGCGAGACCCCUGCUGACACAUAGCCGUCGUCCCGCUCUGGCACAUUCUCAUUCUCGUUCUCGUU ..-...(((((......)))))......(((.(....).)))(((...(((.((((((.(((..(.((((.....)))).)..))).)))))).)))....)))............... ( -33.60) >DroSec_CAF1 7259 119 - 1 CCAUUCCCUCAUACCGUUAAAACCAAUAUGCCGGCAAGCGCAGGUCACUGCGGGAGCGAGACCCCUGCUGACACAUAGCCGCCAUCCCGCUCUGGCACAUUCUCAUUCUCGUUCUCACU .....((..((((..((....))...))))..)).....((((....))))((((((((((.....((((.....)))).((((........))))..........))))))))))... ( -30.20) >DroSim_CAF1 6472 118 - 1 UC-UUCCUUAAUACCGUUUAAACCAAUAUGCCCGAGAGUGCAGGGCACUGCGGGAGCGAGACCCCUGCUGACACAUAGCCGCCAUCCCGCUCUGGCACAUUCUCAUUCUCGUUCUCGCU ..-.........................(((((.........)))))..((((((((((((.....((((.....)))).((((........))))..........)))))))))))). ( -36.20) >DroEre_CAF1 6515 111 - 1 ---UCCCUUAAUAUCGUUAUA-CUAAUACGCCUCAAAUUGCAGGGCAUUGCGUGAGCGAGACCCCUGCUGACA--CAGCCGCCAUCCCGCUCUGGC--AUUCUCAUUCUCGUUCUCGCU ---..................-.......((((.........))))...(((.((((((((.....((((...--)))).((((........))))--........)))))))).))). ( -30.50) >DroYak_CAF1 6613 114 - 1 ---UUCCUUAAUACCGUUAAAGCCAAUAUGCCUCCAAGUGCAGGGCAUUGCGUGAGCGAGACCCCUGCUGACACACAGCCGCCAUCCCGCUCUGGC--AUUCUCAUUCUCGUUCUCGCU ---..................(((....((((.....).))).)))...(((.((((((((.....((((.....)))).((((........))))--........)))))))).))). ( -35.00) >consensus _C_UUCCUUAAUACCGUUAAAACCAAUAUGCCCGAAAGUGCAGGGCACUGCGGGAGCGAGACCCCUGCUGACACAUAGCCGCCAUCCCGCUCUGGCACAUUCUCAUUCUCGUUCUCGCU .............................((((.........))))...((((((((((((.....((((.....)))).((((........))))..........)))))))))))). (-27.46 = -27.26 + -0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:05 2006