
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,004,980 – 12,005,072 |
| Length | 92 |
| Max. P | 0.999947 |

| Location | 12,004,980 – 12,005,072 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 90.20 |
| Mean single sequence MFE | -44.66 |
| Consensus MFE | -39.88 |
| Energy contribution | -40.32 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -4.20 |
| Structure conservation index | 0.89 |
| SVM decision value | 4.76 |
| SVM RNA-class probability | 0.999947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
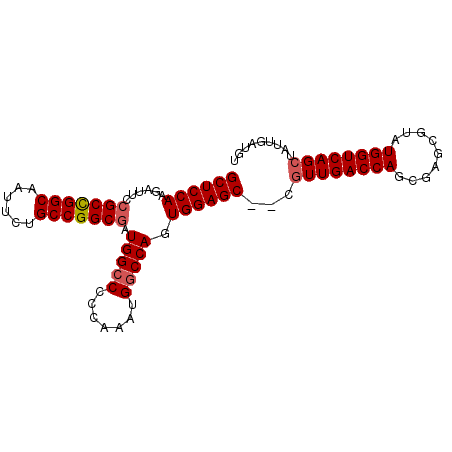
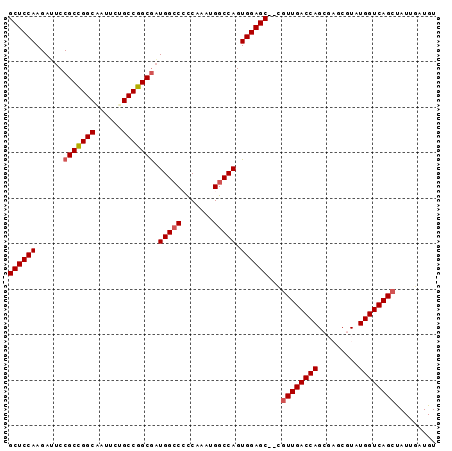
>2L_DroMel_CAF1 12004980 92 + 22407834 GCUCCAAGAUUCCGCCGGCAAUUCUGCCGGCGAUGGCCCCCAAAUGGCCAGUGGAGC--CGUUGACCAGCGAGCGUAUGGUCAGCUAUUGAUGU ((((((......((((((((....)))))))).(((((.......))))).))))))--.((((((((((....)).))))))))......... ( -46.60) >DroSec_CAF1 5760 94 + 1 GCUCCAAGAUUCCGCUGGCAAUUCUGCCGGCGAUGGCCCCCAAAUGGCCAGUGGAGCCCCGUUGACCAGCGAGCGUAUGGUCAGCUAUUGAUGU ((((((......((((((((....)))))))).(((((.......))))).))))))...((((((((((....)).))))))))......... ( -44.40) >DroSim_CAF1 4964 94 + 1 GCUCCAAGAUUCCGCCGGCGAUUCUGCCGGCGAUGGCCCCCAAAUGGCCAGUGGAGCCCCAUUGACCAGCGAGCGUAUGGUCAGCUAUUGAUGU ((((((......((((((((....)))))))).(((((.......))))).))))))....(((((((((....)).))))))).......... ( -42.20) >DroEre_CAF1 4962 87 + 1 GCUCCAAGAGUCCGCCGGCUAUUCUGCCGGCGUUGGCC-CCAAAUGGCCAGUGGAGC--UGUUGACCAG----CGUAUGGUCAGCCAUUGAUGU ((((((......(((((((......)))))))((((((-......))))))))))))--.((((((((.----....))))))))......... ( -46.00) >DroYak_CAF1 5080 87 + 1 GCUCCAAGAGUCCGCCGGCUAUUCUGCCGGCAAUGGGC-UCAAAUGGCCAGUGGAGC--AGUUGACCAG----CGUAUGGUCAGCUAUUGAUGU ((((((.((((((((((((......))))))...))))-))..........))))))--(((((((((.----....)))))))))........ ( -44.10) >consensus GCUCCAAGAUUCCGCCGGCAAUUCUGCCGGCGAUGGCCCCCAAAUGGCCAGUGGAGC__CGUUGACCAGCGAGCGUAUGGUCAGCUAUUGAUGU ((((((......(((((((......))))))).(((((.......))))).))))))...((((((((.........))))))))......... (-39.88 = -40.32 + 0.44)

| Location | 12,004,980 – 12,005,072 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 90.20 |
| Mean single sequence MFE | -38.06 |
| Consensus MFE | -35.76 |
| Energy contribution | -35.84 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.54 |
| SVM RNA-class probability | 0.999918 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12004980 92 - 22407834 ACAUCAAUAGCUGACCAUACGCUCGCUGGUCAACG--GCUCCACUGGCCAUUUGGGGGCCAUCGCCGGCAGAAUUGCCGGCGGAAUCUUGGAGC .........(.((((((.........)))))).).--((((((.(((((.......)))))((((((((......)))))))).....)))))) ( -40.20) >DroSec_CAF1 5760 94 - 1 ACAUCAAUAGCUGACCAUACGCUCGCUGGUCAACGGGGCUCCACUGGCCAUUUGGGGGCCAUCGCCGGCAGAAUUGCCAGCGGAAUCUUGGAGC .........(.((((((.........)))))).)...((((((.(((((.......)))))((((.(((......))).)))).....)))))) ( -34.10) >DroSim_CAF1 4964 94 - 1 ACAUCAAUAGCUGACCAUACGCUCGCUGGUCAAUGGGGCUCCACUGGCCAUUUGGGGGCCAUCGCCGGCAGAAUCGCCGGCGGAAUCUUGGAGC ...........((((((.........)))))).....((((((.(((((.......)))))((((((((......)))))))).....)))))) ( -39.50) >DroEre_CAF1 4962 87 - 1 ACAUCAAUGGCUGACCAUACG----CUGGUCAACA--GCUCCACUGGCCAUUUGG-GGCCAACGCCGGCAGAAUAGCCGGCGGACUCUUGGAGC .......((..((((((....----.)))))).))--((((((.(((((......-))))).(((((((......)))))))......)))))) ( -41.80) >DroYak_CAF1 5080 87 - 1 ACAUCAAUAGCUGACCAUACG----CUGGUCAACU--GCUCCACUGGCCAUUUGA-GCCCAUUGCCGGCAGAAUAGCCGGCGGACUCUUGGAGC ........((.((((((....----.)))))).))--((((((..((((....).-)))..((((((((......)))))))).....)))))) ( -34.70) >consensus ACAUCAAUAGCUGACCAUACGCUCGCUGGUCAACG__GCUCCACUGGCCAUUUGGGGGCCAUCGCCGGCAGAAUAGCCGGCGGAAUCUUGGAGC .........(.((((((.........)))))).)...((((((.(((((.......))))).(((((((......)))))))......)))))) (-35.76 = -35.84 + 0.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:02 2006