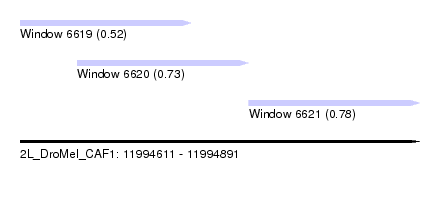
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,994,611 – 11,994,891 |
| Length | 280 |
| Max. P | 0.775037 |

| Location | 11,994,611 – 11,994,731 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.56 |
| Mean single sequence MFE | -36.23 |
| Consensus MFE | -31.05 |
| Energy contribution | -31.72 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.86 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11994611 120 + 22407834 AGCUUGUGCAGCAGGCUGCAGCUGUUGCUGCUGUGGCUGUCCAAAGACACUUUGUGCACCUGUUGCCGCAGCUCCUACACCAAAAAUGCUAUUCCUUGCAUUGGCAUUGCUAAUCGUUCC ((((((.(((((((((..((((.......))))..))((..(((((...)))))..)).))))))))).))))...........(((((((..........)))))))............ ( -38.20) >DroSec_CAF1 6291 111 + 1 AGCUUGAGCAGCAGGCUGCAGCU---------GUGGCUGUCCAAACACACUUGUUGCACCUGUUGCCGCAGCUCCUACACCAAAUAUGCUAUUCCCUGCAUUGGCAUUGCUCAUCGUUCC ((((((.((((((((.((((((.---------(((............)))..)))))))))))))))).))))......(((...((((........)))))))................ ( -34.70) >DroSim_CAF1 6269 120 + 1 AGCUUGAGCAGCAGGCUGCAGCUGUUGCUGCUGUGGCUGUCCAAACACACUUGUUGCACCUGUUGCCGCAGCUCCCACACCAAAUAUGCUAUUCCCUGCAUUGGCAUUGCUCAUCGUUCC ((((((.((((((((..(((((....)))))((..((.((........))..))..)))))))))))).))))......(((...((((........)))))))................ ( -35.80) >consensus AGCUUGAGCAGCAGGCUGCAGCUGUUGCUGCUGUGGCUGUCCAAACACACUUGUUGCACCUGUUGCCGCAGCUCCUACACCAAAUAUGCUAUUCCCUGCAUUGGCAUUGCUCAUCGUUCC ((((((.((((((((.((((((...........(((....))).........)))))))))))))))).))))......(((...((((........)))))))................ (-31.05 = -31.72 + 0.67)

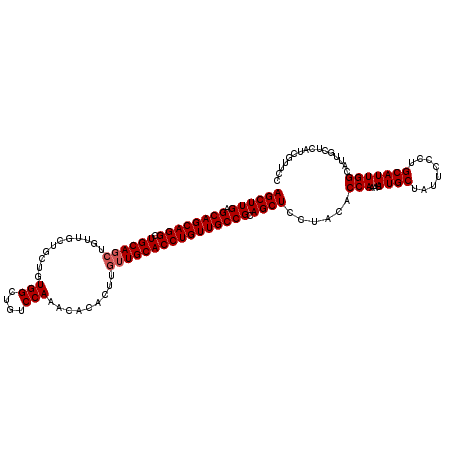
| Location | 11,994,651 – 11,994,771 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -34.70 |
| Consensus MFE | -33.47 |
| Energy contribution | -33.47 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11994651 120 + 22407834 CCAAAGACACUUUGUGCACCUGUUGCCGCAGCUCCUACACCAAAAAUGCUAUUCCUUGCAUUGGCAUUGCUAAUCGUUCCGCCGAAAAUGCUAUUGGUGUUGCUGGCUUGGCUUGUGGCA .(((((...))))).........((((((((((((.(((((((.(((((........)))))((((((.....(((......))).)))))).)))))))....))...))).))))))) ( -35.80) >DroSec_CAF1 6322 120 + 1 CCAAACACACUUGUUGCACCUGUUGCCGCAGCUCCUACACCAAAUAUGCUAUUCCCUGCAUUGGCAUUGCUCAUCGUUCCUCCGAAAAUGCUAUUGGUGUUGCUGGCUUGGCUUGUGGCA ...((((....))))........((((((((((((.(((((((..((((........))))(((((((.....(((......))).))))))))))))))....))...))).))))))) ( -34.20) >DroSim_CAF1 6309 120 + 1 CCAAACACACUUGUUGCACCUGUUGCCGCAGCUCCCACACCAAAUAUGCUAUUCCCUGCAUUGGCAUUGCUCAUCGUUCCUCCGAAAAUGCUAUUGGUGUUGCUGGCUUGGCUUGUGGCA ...........(((..((......((((.((((..((((((((..((((........))))(((((((.....(((......))).))))))))))))).))..)))))))).))..))) ( -34.10) >consensus CCAAACACACUUGUUGCACCUGUUGCCGCAGCUCCUACACCAAAUAUGCUAUUCCCUGCAUUGGCAUUGCUCAUCGUUCCUCCGAAAAUGCUAUUGGUGUUGCUGGCUUGGCUUGUGGCA .......................((((((((((((.(((((((..((((........))))(((((((.....(((......))).))))))))))))))....))...))).))))))) (-33.47 = -33.47 + -0.00)


| Location | 11,994,771 – 11,994,891 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -45.57 |
| Consensus MFE | -42.62 |
| Energy contribution | -43.40 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11994771 120 + 22407834 UUCCCAAAUAUAUUUCCCGCACUCAGCGUGGGCUUUCCAAAAAUGGAUGCAGAUGUAUUGGCGGCAGCCGGAGCUGCUGCAGCGAAGGGAUUACUGGACUGCUGAGGGGCAGCCGACACA .((((.((((((((((((((.....))).))((..((((....)))).))))))))))).((((((((....))))))))......))))....(((.(((((....))))))))..... ( -46.70) >DroSec_CAF1 6442 120 + 1 UUCCCAAAUAUAUUUCCCGCACUCAGUGUGGGCUUUCCAAAAAUGGAUGCAGAUGUAUUGGCGGCGGACGGAGCUGCUGCAGCGAAGGGAUUACUGGGAUGCUGAGGGGCAGCCGACACA .((((.(((((((((.((((((...))))))((..((((....)))).))))))))))).(((((((......)))))))......))))......((.((((....)))).))...... ( -41.70) >DroSim_CAF1 6429 120 + 1 UUCCCAAAUAUAUUUCCCGCACUCAGUGUGGGCUUUCCAAAAAUGGAUGCAGAUGUAUUGGCGGCGGACGGAGCUGCUGCAGCGAAGGGAUUACUGGGCUGCUGAGGGGCAGCCGACACA .((((.(((((((((.((((((...))))))((..((((....)))).))))))))))).(((((((......)))))))......))))......(((((((....)))))))...... ( -48.30) >consensus UUCCCAAAUAUAUUUCCCGCACUCAGUGUGGGCUUUCCAAAAAUGGAUGCAGAUGUAUUGGCGGCGGACGGAGCUGCUGCAGCGAAGGGAUUACUGGGCUGCUGAGGGGCAGCCGACACA .((((.(((((((((.((((((...))))))((..((((....)))).))))))))))).(((((((......)))))))......))))......(((((((....)))))))...... (-42.62 = -43.40 + 0.78)

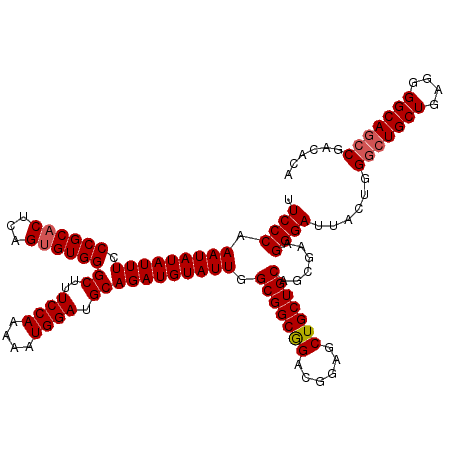
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:58 2006