| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,985,694 – 11,985,801 |
| Length | 107 |
| Max. P | 0.771276 |

| Location | 11,985,694 – 11,985,801 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 81.36 |
| Mean single sequence MFE | -22.36 |
| Consensus MFE | -16.26 |
| Energy contribution | -16.27 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649643 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11985694 107 + 22407834 CCAUCGCCAAAGGCGCAAGAUCCUCGGCUUUAAGUUUCUGCCGAAGUUUCUUAACCUUUUUCUUUGGCUUUUUAAAUCUGGAAUUUAAUAAACGUA--UUUGCAUUAAA .....(((...)))(((((.....((...(((((((((.(((((((...............)))))))...........)))))))))....))..--)))))...... ( -20.56) >DroPse_CAF1 44203 108 + 1 CCUCAGCCAAAGGCUCGAGGUCUUCGGCUUUCAGUUUCUGCCGAAGCUUCUUUACCUUUUUCUUUGGCUUCUUGAAUCUAAAGGG-AAUGAGAUUAUUUUGGAAUGAAG ..(((.((((((.(((((((.(((((((...........))))))))))).......((..((((((.((....)).))))))..-)).)))....))))))..))).. ( -33.20) >DroSec_CAF1 3714 107 + 1 CCAUCGCCAAAGGCUCAAGAUCCUCGGCUUUAAGUUUCUGCCGAAGUUUCUUAACCUUUUUCUUUGGCUUUUUAAAUCUGGAAAUUUAUAAACGUG--UUUGCAUUAAA .....((((((((...((((..((((((...........)))).))..))))........))))))))..........((.((((..........)--))).))..... ( -20.80) >DroEre_CAF1 3713 107 + 1 CUUCCGCCAAAGGCUCAAGAUCCUCGGCUUUUAGUUUCUGUCGAAGUUUCUUAACCUUCUUCUUUGGCUUCUUAAAUCUGGAAAUAAAUAAAAGUA--UUUGCAUUAAA .....((((((((...((((..((((((...........)))).))..))))........))))))))..........((.(((((........))--))).))..... ( -19.50) >DroYak_CAF1 3740 107 + 1 CCUCUGCCAAAGGCUCAAGAUCCUCGGCUUUUAGUUUCUGCCGAAGUUUCUUAACCUUUUUCUUUGGCUUUUUAAAUCUGUAAAUAAAAAAACGCA--UUUGCAUUAAA .....((((((((...((((..((((((...........)))).))..))))........))))))))..........(((((((..........)--))))))..... ( -22.20) >DroAna_CAF1 35569 99 + 1 CCUCCGGCAAAGGCUCCAGAUCCUCGGCCUUCAGUUUCUGGCGCAGUUUCUUGACCUUCUUCUUUGGCUUUUUAAACCUGAAAC-AUAAAAUCG---------AUUAAU .....(.(((((((.(((((..((........))..))))).)).(((....)))......))))).)................-.........---------...... ( -17.90) >consensus CCUCCGCCAAAGGCUCAAGAUCCUCGGCUUUAAGUUUCUGCCGAAGUUUCUUAACCUUUUUCUUUGGCUUUUUAAAUCUGGAAAUAAAUAAACGUA__UUUGCAUUAAA .....((((((((...((((..((((((...........)))).))..))))........))))))))......................................... (-16.26 = -16.27 + 0.00)

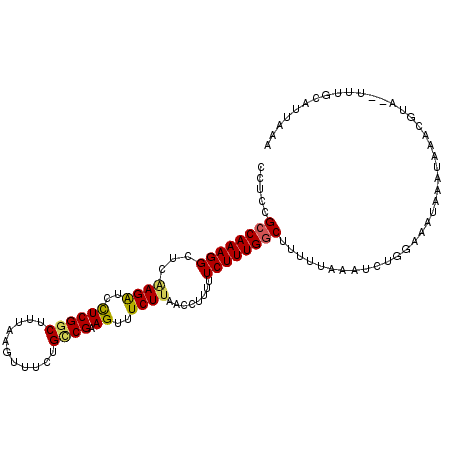
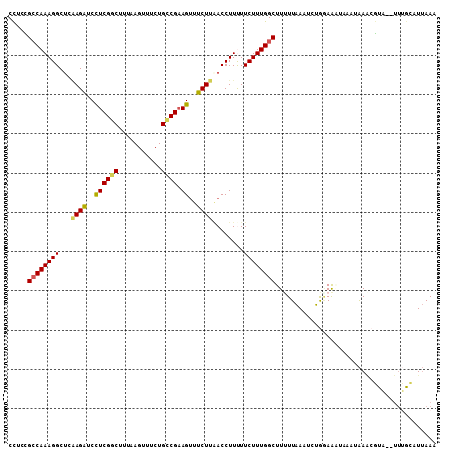
| Location | 11,985,694 – 11,985,801 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 81.36 |
| Mean single sequence MFE | -24.68 |
| Consensus MFE | -18.03 |
| Energy contribution | -19.28 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
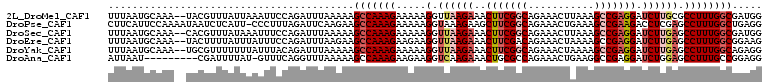
>2L_DroMel_CAF1 11985694 107 - 22407834 UUUAAUGCAAA--UACGUUUAUUAAAUUCCAGAUUUAAAAAGCCAAAGAAAAAGGUUAAGAAACUUCGGCAGAAACUUAAAGCCGAGGAUCUUGCGCCUUUGGCGAUGG .....(((...--...((((.(((((((...))))))).)))).......(((((((((((..(((((((...........))))))).))))).)))))).))).... ( -25.30) >DroPse_CAF1 44203 108 - 1 CUUCAUUCCAAAAUAAUCUCAUU-CCCUUUAGAUUCAAGAAGCCAAAGAAAAAGGUAAAGAAGCUUCGGCAGAAACUGAAAGCCGAAGACCUCGAGCCUUUGGCUGAGG ((((..........(((((.(..-....).))))).....((((((((....((((.......(((((((...........))))))))))).....)))))))))))) ( -26.11) >DroSec_CAF1 3714 107 - 1 UUUAAUGCAAA--CACGUUUAUAAAUUUCCAGAUUUAAAAAGCCAAAGAAAAAGGUUAAGAAACUUCGGCAGAAACUUAAAGCCGAGGAUCUUGAGCCUUUGGCGAUGG ...........--..((((..((((((....))))))....(((((((.....(.((((((..(((((((...........))))))).)))))).)))))))))))). ( -27.50) >DroEre_CAF1 3713 107 - 1 UUUAAUGCAAA--UACUUUUAUUUAUUUCCAGAUUUAAGAAGCCAAAGAAGAAGGUUAAGAAACUUCGACAGAAACUAAAAGCCGAGGAUCUUGAGCCUUUGGCGGAAG ...........--..(((..((((......))))..)))..(((((((.....(.((((((..(((((...............))))).)))))).))))))))..... ( -19.56) >DroYak_CAF1 3740 107 - 1 UUUAAUGCAAA--UGCGUUUUUUUAUUUACAGAUUUAAAAAGCCAAAGAAAAAGGUUAAGAAACUUCGGCAGAAACUAAAAGCCGAGGAUCUUGAGCCUUUGGCAGAGG ...(((((...--.)))))......................(((((((.....(.((((((..(((((((...........))))))).)))))).))))))))..... ( -28.30) >DroAna_CAF1 35569 99 - 1 AUUAAU---------CGAUUUUAU-GUUUCAGGUUUAAAAAGCCAAAGAAGAAGGUCAAGAAACUGCGCCAGAAACUGAAGGCCGAGGAUCUGGAGCCUUUGCCGGAGG .....(---------(...((((.-(((((.((((.....)))).........(((((......)).))).)))))))))(((.((((........)))).))).)).. ( -21.30) >consensus UUUAAUGCAAA__UACGUUUAUUUAUUUCCAGAUUUAAAAAGCCAAAGAAAAAGGUUAAGAAACUUCGGCAGAAACUAAAAGCCGAGGAUCUUGAGCCUUUGGCGGAGG .........................................(((((((.....(.((((((..(((((((...........))))))).)))))).))))))))..... (-18.03 = -19.28 + 1.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:54 2006