
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,980,307 – 11,980,437 |
| Length | 130 |
| Max. P | 0.999019 |

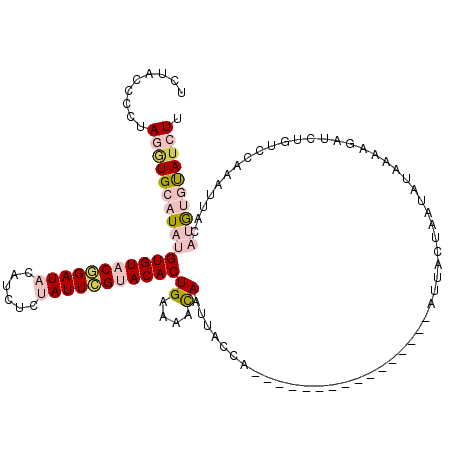
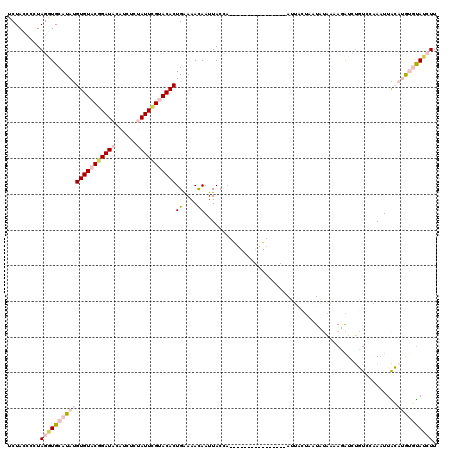
| Location | 11,980,307 – 11,980,411 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.94 |
| Mean single sequence MFE | -20.87 |
| Consensus MFE | -11.34 |
| Energy contribution | -13.54 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.54 |
| SVM decision value | 3.33 |
| SVM RNA-class probability | 0.999019 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11980307 104 - 22407834 UCUACCCCUAGAUGCAUAUGUGUACGGAUACAUCUCUAUUCGUACACUGAAAACAAUUACCA----------------AUUCCUAUUAUAAUAGAUCUGUCUAAAUUACAUGUGUAUCUU .........(((((((((((((((((((((......))))))))).................----------------.............((((....))))....)))))))))))). ( -25.80) >DroSec_CAF1 199715 104 - 1 UCUACCCCUAGGUGCAUAUGUGUACGGAUACAUCUCUAUUCGUACACUGAAAACAAUUACCA----------------AUUACUAAUAUAAAAUAUCUGUCCAAAUUACAUGUGUAUCUU .........(((((((((((((((((((((......))))))))).................----------------...((..((((...))))..)).......)))))))))))). ( -22.40) >DroSim_CAF1 166171 104 - 1 UCUACCCCUAGGUGCAUAUGUGUACGGAUACAUCUCUAUUCGUACACUGGAAACAAUUACCA----------------AUUACUAAUAUAAAAUAUCUGUCCAAAUUACAUGUGUAUCUU .........(((((((((((((((((((((......)))))))))..((....)).......----------------.............................)))))))))))). ( -26.10) >DroEre_CAF1 174306 119 - 1 UCUACCCCUAGGUGCAUACGUGUACAGAUAUGUCUUCAUUCGCACACUGGAAAUAAUUACGUUGCGCCUCGUAUUUCAAAAACUUUUCUAUAAGAUCUUUUCUAUUUGAUCAACUAU-UU .........(((((((.(((((..(((...(((........)))..)))........))))))))))))........................((((..........))))......-.. ( -18.70) >DroYak_CAF1 171338 114 - 1 UCUACCCCUAGUUGCAUAUGUGUACAGAUAUAUCUUCAUUUGCACACUGAAAAUAAUUACUAUAUGUCAUGUAUUUAAAAG-----UUAGUAAAGUACUUAACAAUUGAUCAAUCAU-UU ........((((((.....((((.(((((........))))).)))).........((((((...................-----.)))))).........)))))).........-.. ( -11.35) >consensus UCUACCCCUAGGUGCAUAUGUGUACGGAUACAUCUCUAUUCGUACACUGAAAACAAUUACCA________________AUUACUAAUAUAAAAGAUCUGUCCAAAUUACAUGUGUAUCUU .........(((((((((((((((((((((......)))))))))))((....))......................................................)))))))))). (-11.34 = -13.54 + 2.20)

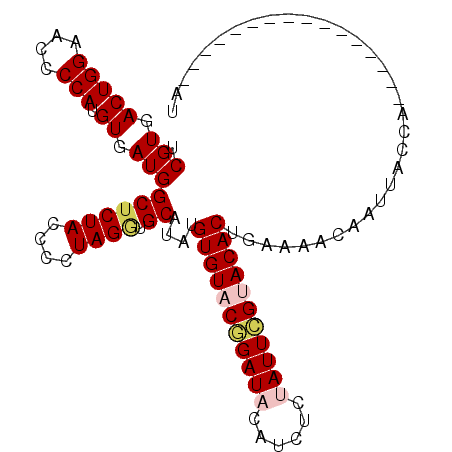
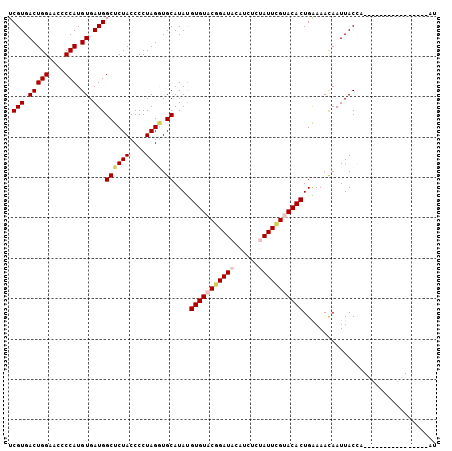
| Location | 11,980,347 – 11,980,437 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 82.21 |
| Mean single sequence MFE | -22.86 |
| Consensus MFE | -18.56 |
| Energy contribution | -19.40 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.605974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11980347 90 - 22407834 UCGUGACUGGAACCCCAUGUGAUGGCUCUACCCCUAGAUGCAUAUGUGUACGGAUACAUCUCUAUUCGUACACUGAAAACAAUUACCA----------------AU .(((.(((((....))).)).)))((((((....)))).))....(((((((((((......)))))))))))...............----------------.. ( -23.80) >DroSec_CAF1 199755 90 - 1 UCGUGACUGGAACCCCAUGUGAUGGCUCUACCCCUAGGUGCAUAUGUGUACGGAUACAUCUCUAUUCGUACACUGAAAACAAUUACCA----------------AU .(((.(((((....))).)).)))((((((....)))).))....(((((((((((......)))))))))))...............----------------.. ( -22.90) >DroSim_CAF1 166211 90 - 1 UCGUGACUGGAACCCCAUGUGAUGGCUCUACCCCUAGGUGCAUAUGUGUACGGAUACAUCUCUAUUCGUACACUGGAAACAAUUACCA----------------AU ((((.(((((....))).)).))))...........((((.....(((((((((((......)))))))))))((....))..)))).----------------.. ( -26.40) >DroEre_CAF1 174345 106 - 1 UCGUGACUGGAACCCCAUGUGAUGGCUCUACCCCUAGGUGCAUACGUGUACAGAUAUGUCUUCAUUCGCACACUGGAAAUAAUUACGUUGCGCCUCGUAUUUCAAA ((((.(((((....))).)).))))..........(((((((.(((((..(((...(((........)))..)))........))))))))))))........... ( -21.90) >DroYak_CAF1 171372 106 - 1 UCGUGACUGGAACCCCAUGUGAUGGCUCUACCCCUAGUUGCAUAUGUGUACAGAUAUAUCUUCAUUUGCACACUGAAAAUAAUUACUAUAUGUCAUGUAUUUAAAA .(((((((((....))).(((((.((.(((....)))..))....((((.(((((........))))).))))........))))).....))))))......... ( -19.30) >consensus UCGUGACUGGAACCCCAUGUGAUGGCUCUACCCCUAGGUGCAUAUGUGUACGGAUACAUCUCUAUUCGUACACUGAAAACAAUUACCA________________AU .(((.(((((....))).)).)))((((((....)))).))....(((((((((((......)))))))))))................................. (-18.56 = -19.40 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:50 2006