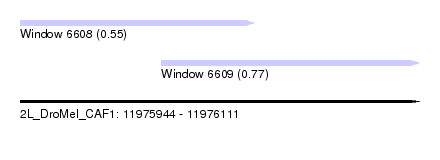
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,975,944 – 11,976,111 |
| Length | 167 |
| Max. P | 0.769765 |

| Location | 11,975,944 – 11,976,042 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 78.43 |
| Mean single sequence MFE | -28.72 |
| Consensus MFE | -16.12 |
| Energy contribution | -16.23 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.553407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
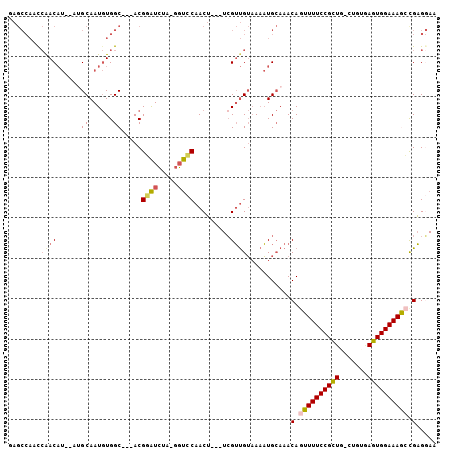
>2L_DroMel_CAF1 11975944 98 + 22407834 GAGCCAACCGACAUGCAUGCAAUGUGGC---ACGGAUCUA-GGUCCAACU---UCGUUGUAAAAUGCAAACAGUUUUCCGCUG-UUGUGAGUGGAAAGCUGAGGAA .......((....(((((((((((.((.---..((((...-.))))..))---.))))))...)))))..((((((((((((.-.....)))))))))))).)).. ( -35.20) >DroSec_CAF1 195385 98 + 1 GAGCCAACCGACAUGCAUGCAAUGUGGC---ACGGAUCUA-GGUCCAACU---UCGUUGUAAAAUGCAAACAGUUUUCCGCUG-UUGAGAGUGGAAAGCCGAGGAA .......((....(((((((((((.((.---..((((...-.))))..))---.))))))...)))))..(.((((((((((.-.....)))))))))).).)).. ( -30.60) >DroSim_CAF1 161773 98 + 1 GAGCCAACCGACAUGCAUGCAAUGUGGC---ACGGAUCUA-GGUCCAACU---UCGUUGUAAAAUGCAAACAGUUUUCCGCUG-UUGAGAGUGGAAAGCCGAGGAA .......((....(((((((((((.((.---..((((...-.))))..))---.))))))...)))))..(.((((((((((.-.....)))))))))).).)).. ( -30.60) >DroEre_CAF1 169816 94 + 1 AAGCCAUCCAACAU----GCAAUGUGGC---ACGGAUCUA-GGUACAACU---UGGUUGUAAAAUGCAAACAGUUUUCCGCUG-CUGUGAGUGGAAAGCCGAGCAA ..(((((((....(----((......))---).))))...-)))(((((.---..)))))....(((...(.((((((((((.-.....)))))))))).).))). ( -23.90) >DroYak_CAF1 166784 94 + 1 GAGCCAUUCAACAU----GCAAUGUGGC---ACGGAUCUG-GGUCCAACU---UCGUUGUAAAAUGCAAACAGUUUUCCGCUG-CUGUCAGUGGAAAAGCGAGGAA ..(((((.......----.....)))))---..((((...-.))))..((---(((((((.....))).....((((((((((-....)))))))))))))))).. ( -30.20) >DroMoj_CAF1 187403 93 + 1 -------CAAAC----AUGCAAUAUGGCUGAACGGGUGGCUGCCUCAACACAGUCGACGGCACUUGAAAACA-UUUUCCGUUGCCUGCAAAUGGAAAAUUGUG-AA -------.....----.(((....((((((...((((....)))).....))))))...))).......(((-(((((((((.......))))))))).))).-.. ( -21.80) >consensus GAGCCAACCAACAU__AUGCAAUGUGGC___ACGGAUCUA_GGUCCAACU___UCGUUGUAAAAUGCAAACAGUUUUCCGCUG_CUGUGAGUGGAAAGCCGAGGAA ........((((......((......)).....((((.....)))).........))))...........(.((((((((((.......)))))))))).)..... (-16.12 = -16.23 + 0.11)


| Location | 11,976,003 – 11,976,111 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.00 |
| Mean single sequence MFE | -29.88 |
| Consensus MFE | -13.62 |
| Energy contribution | -13.43 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.769765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11976003 108 + 22407834 CAAACAGUUUUCCGCUG-UUGUGAGUGGAAAGCUGAGGAAAAAUUAUGUGCUAAACACGCAGUCUGA-G----CG-AGUACAGUUAGCACAAAA--UAU-AAAGAACAUUCUCGUCCA-- ....((((((((((((.-.....)))))))))))).(((...(((.((((((((((.(((.......-)----))-.))....)))))))).))--)..-...((......)).))).-- ( -34.10) >DroVir_CAF1 177721 116 + 1 CAAACA-UUUUCCGCUGCCUGCAAAUGGAAAAUUGUG-AAAAAUGAUGUGCUAAACACGAAGCUUGCAACGGCUGUGGUACAGUUACUGCAAAAGGC--UGUACUCCAUUCUUCGCUUCC .....(-(((((((.((....))..)))))))).(((-((.((((..((((.........((((((((..(((((.....)))))..))))...)))--)))))..)))).))))).... ( -28.70) >DroSim_CAF1 161832 108 + 1 CAAACAGUUUUCCGCUG-UUGAGAGUGGAAAGCCGAGGAAAAAUUAUGUGCUAAACACGCAGUCUGA-G----CG-AGUACAGUUAGCACAAAA--UAU-AAAGCACAUUCUCGUCUA-- ......((((((((((.-.....))))))))))((((((...(((.((((((((((.(((.......-)----))-.))....)))))))).))--)..-........))))))....-- ( -29.32) >DroEre_CAF1 169871 108 + 1 CAAACAGUUUUCCGCUG-CUGUGAGUGGAAAGCCGAGCAAAAAUGAUGCGCUAAACACGCAGGCUGA-G----UG-AGUACAGUUAGCACAAGC--UAU-AAAGCACAUUCCCGUCUA-- ....(.((((((((((.-.....)))))))))).).(((.......)))((.......))((((.((-(----((-.((.....((((....))--)).-...)).)))))..)))).-- ( -27.90) >DroYak_CAF1 166839 110 + 1 CAAACAGUUUUCCGCUG-CUGUCAGUGGAAAAGCGAGGAAAAAUUAUGUGCUAAACACGCAGUCUGA-G----UG-GGUACAGUUAGCGCAAAAUAUAU-AAAGCACAUUCCCGUCUA-- .......((((((((((-....))))))))))(((.(((......(((((((......((.(.((((-.----((-....)).))))))).........-..)))))))))))))...-- ( -28.53) >DroMoj_CAF1 187458 117 + 1 AAAACA-UUUUCCGUUGCCUGCAAAUGGAAAAUUGUG-AAAAGCUAUGUGCUAAACACGAAGCAUGCAACAGCUG-GGUACAGUUACUGCAAAAAGUCUCGAGCCACAUUCUUAGCUCAC ...(((-(((((((((.......))))))))).))).-....(((.((((.....)))).))).((((..(((((-....)))))..)))).........((((..........)))).. ( -30.70) >consensus CAAACAGUUUUCCGCUG_CUGUGAGUGGAAAACCGAGGAAAAAUUAUGUGCUAAACACGCAGUCUGA_G____UG_AGUACAGUUAGCACAAAA__UAU_AAAGCACAUUCUCGUCUA__ .......(((((((((.......)))))))))..............((((((((..((...................))....))))))))............................. (-13.62 = -13.43 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:46 2006