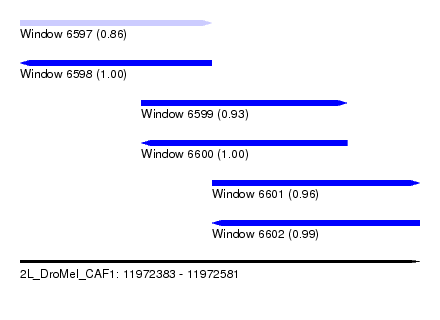
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,972,383 – 11,972,581 |
| Length | 198 |
| Max. P | 0.999782 |

| Location | 11,972,383 – 11,972,478 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 90.42 |
| Mean single sequence MFE | -25.66 |
| Consensus MFE | -19.54 |
| Energy contribution | -20.82 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
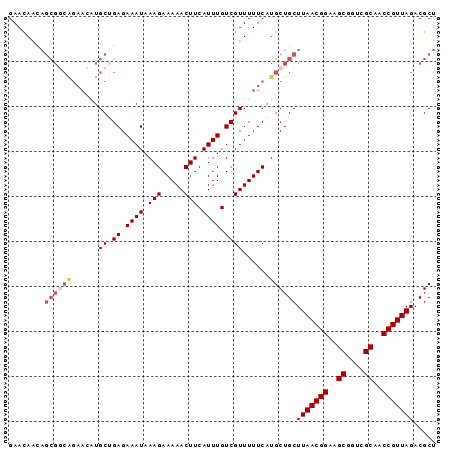
>2L_DroMel_CAF1 11972383 95 + 22407834 GAAUAACAGCGGCAGAACAUGCUGAGAAAUAAAGAAAAACUUCAUUUGUCGUUUUUCAUGCUGCUUAACGGAAGCGGUCGCAACCGUUAGACGCU .......((((((((..((((..(((..((((((((....))).)))))..)))..))))))))(((((((..((....))..))))))).)))) ( -27.00) >DroSec_CAF1 191760 95 + 1 GAACAAAAGCGGCAGAACAUGCUGAGAAAUAAAGAAAAACUUCAUUUGUCGUUUUUCAUGCUGCUUAACGGAAGCGGUCGCAACCGUUAGACGCU .......((((((((..((((..(((..((((((((....))).)))))..)))..))))))))(((((((..((....))..))))))).)))) ( -27.00) >DroSim_CAF1 158122 95 + 1 GAACAACAGCGGCAGAACAUGCUGAGAAAUAAAGAAAAACUUCAUUUGUCGUUUUUCAUGCUGCUUAACGGAAGCGGUCGCAACCGUUAGACGCU .......((((((((..((((..(((..((((((((....))).)))))..)))..))))))))(((((((..((....))..))))))).)))) ( -27.00) >DroEre_CAF1 166455 95 + 1 GAACAACAGCGUCGGAAAGUGCAGAGAAAUAAAGAAAAACUUCAUUUGUCGUUUUUCAUGCUGCUUAACGGAAGCGGUCGCCACCGUUAGACGCU .......((((((.......(((((....)...(((((((..(....)..)))))))...)))).((((((..((....))..)))))))))))) ( -28.20) >DroYak_CAF1 163214 81 + 1 GAACA--------------UGCAGAGAAAUAAAGAAAAACUUCAUUUGUCGUUUUUCAUGCUGCUUAACGGAAGCGUUCGCCACCGUUAGACGCU .....--------------.(((((....)...(((((((..(....)..)))))))...))))(((((((..((....))..)))))))..... ( -19.10) >consensus GAACAACAGCGGCAGAACAUGCUGAGAAAUAAAGAAAAACUUCAUUUGUCGUUUUUCAUGCUGCUUAACGGAAGCGGUCGCAACCGUUAGACGCU ........((((((......((.((.((((.(((.....))).)))).))))......))))))(((((((..((....))..)))))))..... (-19.54 = -20.82 + 1.28)

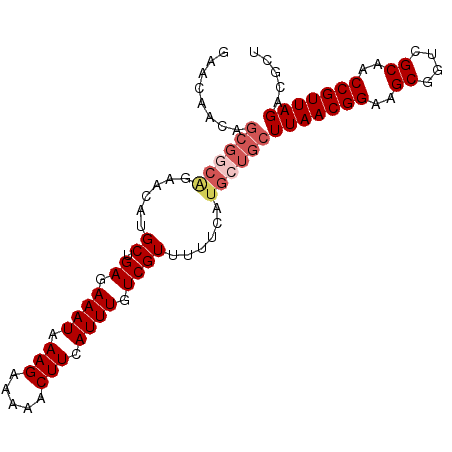
| Location | 11,972,383 – 11,972,478 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 90.42 |
| Mean single sequence MFE | -26.76 |
| Consensus MFE | -22.26 |
| Energy contribution | -22.26 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.996561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11972383 95 - 22407834 AGCGUCUAACGGUUGCGACCGCUUCCGUUAAGCAGCAUGAAAAACGACAAAUGAAGUUUUUCUUUAUUUCUCAGCAUGUUCUGCCGCUGUUAUUC ((((..((((((..((....))..)))))).((((((((......((.((((((((.....)))))))).))..))))..))))))))....... ( -26.80) >DroSec_CAF1 191760 95 - 1 AGCGUCUAACGGUUGCGACCGCUUCCGUUAAGCAGCAUGAAAAACGACAAAUGAAGUUUUUCUUUAUUUCUCAGCAUGUUCUGCCGCUUUUGUUC ((((..((((((..((....))..)))))).((((((((......((.((((((((.....)))))))).))..))))..))))))))....... ( -26.20) >DroSim_CAF1 158122 95 - 1 AGCGUCUAACGGUUGCGACCGCUUCCGUUAAGCAGCAUGAAAAACGACAAAUGAAGUUUUUCUUUAUUUCUCAGCAUGUUCUGCCGCUGUUGUUC ((((..((((((..((....))..)))))).((((((((......((.((((((((.....)))))))).))..))))..))))))))....... ( -26.80) >DroEre_CAF1 166455 95 - 1 AGCGUCUAACGGUGGCGACCGCUUCCGUUAAGCAGCAUGAAAAACGACAAAUGAAGUUUUUCUUUAUUUCUCUGCACUUUCCGACGCUGUUGUUC ((((((((((((.(((....))).)))))).((((...(((((((..(....)..))))))).........)))).......))))))....... ( -30.00) >DroYak_CAF1 163214 81 - 1 AGCGUCUAACGGUGGCGAACGCUUCCGUUAAGCAGCAUGAAAAACGACAAAUGAAGUUUUUCUUUAUUUCUCUGCA--------------UGUUC ...(..((((((.(((....))).))))))..)((((((......((.((((((((.....)))))))).))..))--------------)))). ( -24.00) >consensus AGCGUCUAACGGUUGCGACCGCUUCCGUUAAGCAGCAUGAAAAACGACAAAUGAAGUUUUUCUUUAUUUCUCAGCAUGUUCUGCCGCUGUUGUUC .(((..((((((..((....))..))))))..)............((.((((((((.....)))))))).)).)).................... (-22.26 = -22.26 + -0.00)

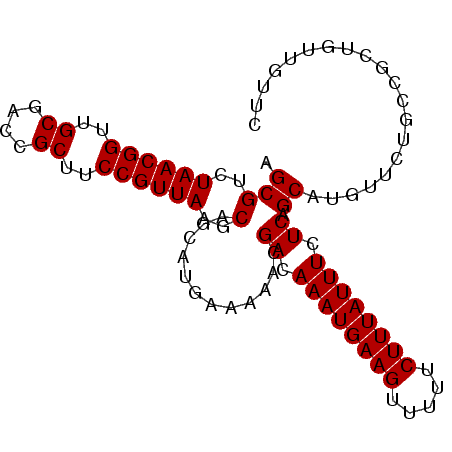
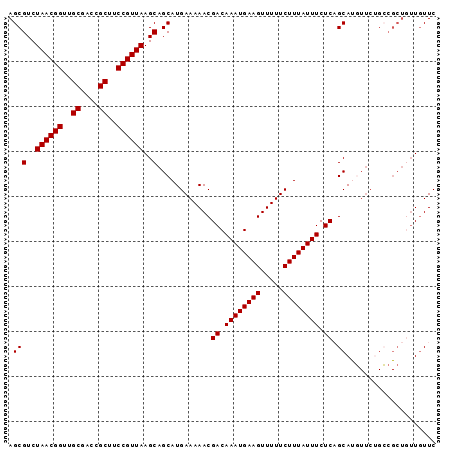
| Location | 11,972,443 – 11,972,545 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 90.88 |
| Mean single sequence MFE | -32.96 |
| Consensus MFE | -25.28 |
| Energy contribution | -25.68 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934628 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11972443 102 + 22407834 CUGCUUAACGGAAGCGGUCGCAACCGUUAGACGCUGCAUCCAAAGCUAUA----------UGCAGCUAUAGCUGCAUCUGAAUCCCAGCUCUCGACGGCGAUCCCAACCAAA .........((..(.((((((...(((((((.((((.......((....(----------((((((....)))))))))......))))))).)))))))))).)..))... ( -31.12) >DroSec_CAF1 191820 102 + 1 CUGCUUAACGGAAGCGGUCGCAACCGUUAGACGCUGCAUCCAAAGCUAUA----------UGCAGCUAUAGCUGCAUCUGAAUCCCAACUCUUGCCGGCGAUCCCAACCAAA ....(((((((..((....))..))))))).(((((((...........(----------((((((....)))))))..((........)).)).)))))............ ( -27.30) >DroSim_CAF1 158182 102 + 1 CUGCUUAACGGAAGCGGUCGCAACCGUUAGACGCUGCAUCCAAAGCUAUA----------UGCAGCUAUAGCUGCAUCUGAAUCCCAACUCUUGCCGGCGAUCCCAACCAAA ....(((((((..((....))..))))))).(((((((...........(----------((((((....)))))))..((........)).)).)))))............ ( -27.30) >DroEre_CAF1 166515 112 + 1 CUGCUUAACGGAAGCGGUCGCCACCGUUAGACGCUGGAUCCAAAGCUAUAUGCAGCUAUAUGCAGCUAUAGCUGCAUCUGAAUCCCAGCUCUGGCCGGCGAUCCCGGCCAAA ..(..((((((..((....))..))))))..)(((((......((....(((((((((((......))))))))))))).....)))))..(((((((.....))))))).. ( -45.80) >DroYak_CAF1 163260 107 + 1 CUGCUUAACGGAAGCGUUCGCCACCGUUAGACGCUGGAUCCAAAGCUAUA-----CUCUAAGCAGCUAUAGCUGCAUCUGAAUCCCAGCUCUUGCCGGCGAUCCCAGCCAAA ..(..((((((..((....))..))))))..)(((((.............-----......(((((....)))))......(((((.((....)).)).))).))))).... ( -33.30) >consensus CUGCUUAACGGAAGCGGUCGCAACCGUUAGACGCUGCAUCCAAAGCUAUA__________UGCAGCUAUAGCUGCAUCUGAAUCCCAGCUCUUGCCGGCGAUCCCAACCAAA ....(((((((..((....))..))))))).(((((.........................(((((....)))))..(((.....))).......)))))............ (-25.28 = -25.68 + 0.40)

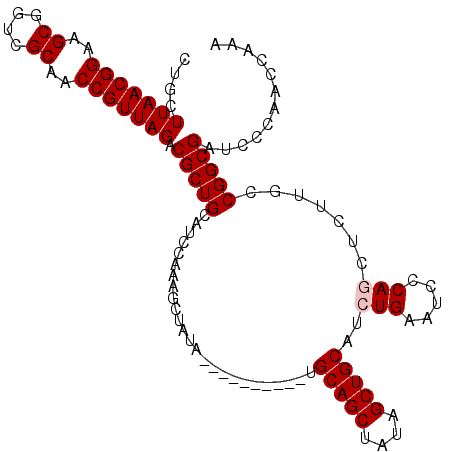
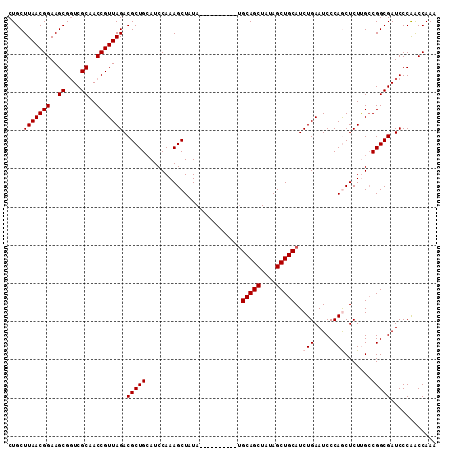
| Location | 11,972,443 – 11,972,545 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 90.88 |
| Mean single sequence MFE | -43.78 |
| Consensus MFE | -37.74 |
| Energy contribution | -37.74 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.86 |
| SVM decision value | 4.07 |
| SVM RNA-class probability | 0.999782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11972443 102 - 22407834 UUUGGUUGGGAUCGCCGUCGAGAGCUGGGAUUCAGAUGCAGCUAUAGCUGCA----------UAUAGCUUUGGAUGCAGCGUCUAACGGUUGCGACCGCUUCCGUUAAGCAG ...(((((.((((((((((.(((((((........(((((((....))))))----------).))))))).))))..))(.....))))).)))))((((.....)))).. ( -38.60) >DroSec_CAF1 191820 102 - 1 UUUGGUUGGGAUCGCCGGCAAGAGUUGGGAUUCAGAUGCAGCUAUAGCUGCA----------UAUAGCUUUGGAUGCAGCGUCUAACGGUUGCGACCGCUUCCGUUAAGCAG ....(((((((((.(((((....))))))))))..(((((((....))))))----------).............))))(..((((((..((....))..))))))..).. ( -38.30) >DroSim_CAF1 158182 102 - 1 UUUGGUUGGGAUCGCCGGCAAGAGUUGGGAUUCAGAUGCAGCUAUAGCUGCA----------UAUAGCUUUGGAUGCAGCGUCUAACGGUUGCGACCGCUUCCGUUAAGCAG ....(((((((((.(((((....))))))))))..(((((((....))))))----------).............))))(..((((((..((....))..))))))..).. ( -38.30) >DroEre_CAF1 166515 112 - 1 UUUGGCCGGGAUCGCCGGCCAGAGCUGGGAUUCAGAUGCAGCUAUAGCUGCAUAUAGCUGCAUAUAGCUUUGGAUCCAGCGUCUAACGGUGGCGACCGCUUCCGUUAAGCAG (((((((((.....)))))))))(((((.(((((((((((((((((......))))))))))......))))))))))))(..((((((.(((....))).))))))..).. ( -58.40) >DroYak_CAF1 163260 107 - 1 UUUGGCUGGGAUCGCCGGCAAGAGCUGGGAUUCAGAUGCAGCUAUAGCUGCUUAGAG-----UAUAGCUUUGGAUCCAGCGUCUAACGGUGGCGAACGCUUCCGUUAAGCAG ....(((((((((.(((((....)))))))))).((((((((....)))))((((((-----.....)))))))))))))(..((((((.(((....))).))))))..).. ( -45.30) >consensus UUUGGUUGGGAUCGCCGGCAAGAGCUGGGAUUCAGAUGCAGCUAUAGCUGCA__________UAUAGCUUUGGAUGCAGCGUCUAACGGUUGCGACCGCUUCCGUUAAGCAG ....(((((((((.(((((....))))))))))....(((((....))))).........................))))(..((((((..((....))..))))))..).. (-37.74 = -37.74 + -0.00)

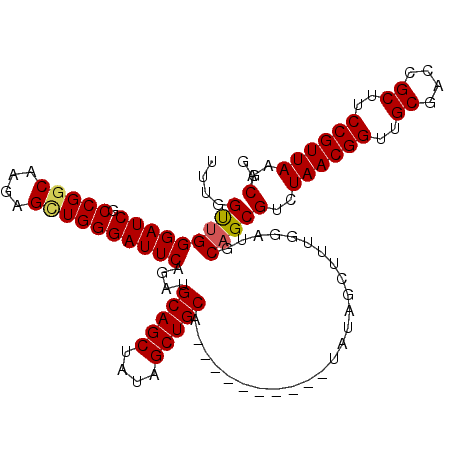
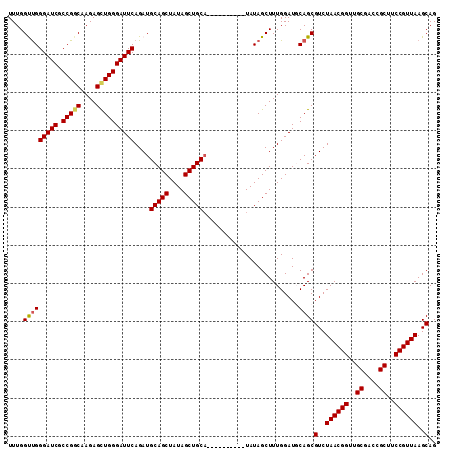
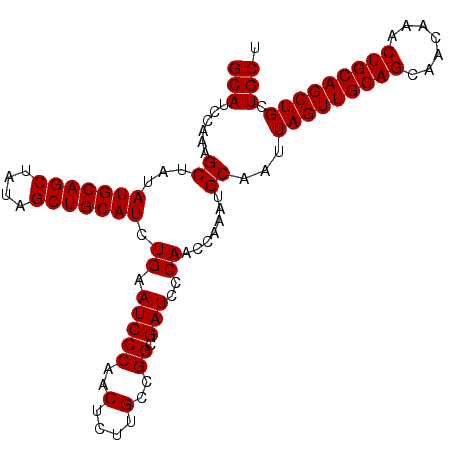
| Location | 11,972,478 – 11,972,581 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 98.06 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -26.93 |
| Energy contribution | -26.93 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11972478 103 + 22407834 GCAUCCAAAGCUAUAUGCAGCUAUAGCUGCAUCUGAAUCCCAGCUCUCGACGGCGAUCCCAACCAAAUGCAAUUAGUUGCAGCAACAAACUGCAGCUGCUGCU .........(((..(((((((....)))))))(((.....)))........)))..............(((..(((((((((.......))))))))).))). ( -28.10) >DroSec_CAF1 191855 103 + 1 GCAUCCAAAGCUAUAUGCAGCUAUAGCUGCAUCUGAAUCCCAACUCUUGCCGGCGAUCCCAACCAAAUGCAAUUAGUUGCAGCAACAAACUGCAGCUGCUGCU (((......((...(((((((....))))))).((.(((((..........)).)))..)).......))...(((((((((.......))))))))).))). ( -26.90) >DroSim_CAF1 158217 103 + 1 GCAUCCAAAGCUAUAUGCAGCUAUAGCUGCAUCUGAAUCCCAACUCUUGCCGGCGAUCCCAACCAAAUGCAAUUAGUUGCAGCAACAAACUGCAGCUGCUGCU (((......((...(((((((....))))))).((.(((((..........)).)))..)).......))...(((((((((.......))))))))).))). ( -26.90) >consensus GCAUCCAAAGCUAUAUGCAGCUAUAGCUGCAUCUGAAUCCCAACUCUUGCCGGCGAUCCCAACCAAAUGCAAUUAGUUGCAGCAACAAACUGCAGCUGCUGCU (((......((...(((((((....))))))).((.(((((..(....)..)).)))..)).......))...(((((((((.......))))))))).))). (-26.93 = -26.93 + 0.00)

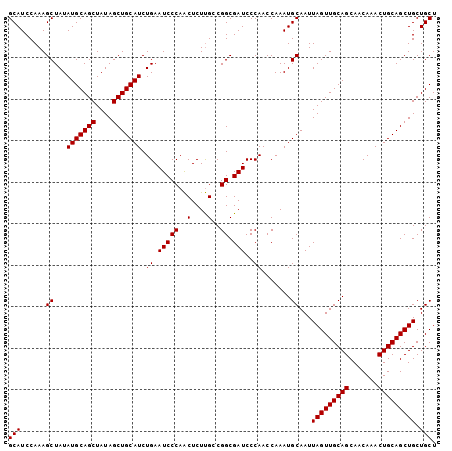
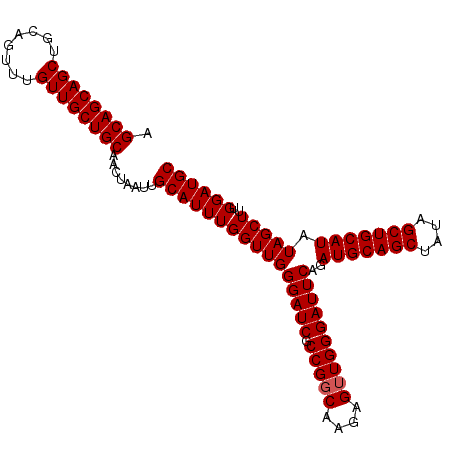
| Location | 11,972,478 – 11,972,581 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 98.06 |
| Mean single sequence MFE | -42.83 |
| Consensus MFE | -42.87 |
| Energy contribution | -43.20 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.27 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11972478 103 - 22407834 AGCAGCAGCUGCAGUUUGUUGCUGCAACUAAUUGCAUUUGGUUGGGAUCGCCGUCGAGAGCUGGGAUUCAGAUGCAGCUAUAGCUGCAUAUAGCUUUGGAUGC .((((((((........))))))))........((((((((((((((((.(((.(....).))))))))..(((((((....))))))).)))))..)))))) ( -41.90) >DroSec_CAF1 191855 103 - 1 AGCAGCAGCUGCAGUUUGUUGCUGCAACUAAUUGCAUUUGGUUGGGAUCGCCGGCAAGAGUUGGGAUUCAGAUGCAGCUAUAGCUGCAUAUAGCUUUGGAUGC .((((((((........))))))))........((((((((((((((((.(((((....))))))))))..(((((((....))))))).)))))..)))))) ( -43.30) >DroSim_CAF1 158217 103 - 1 AGCAGCAGCUGCAGUUUGUUGCUGCAACUAAUUGCAUUUGGUUGGGAUCGCCGGCAAGAGUUGGGAUUCAGAUGCAGCUAUAGCUGCAUAUAGCUUUGGAUGC .((((((((........))))))))........((((((((((((((((.(((((....))))))))))..(((((((....))))))).)))))..)))))) ( -43.30) >consensus AGCAGCAGCUGCAGUUUGUUGCUGCAACUAAUUGCAUUUGGUUGGGAUCGCCGGCAAGAGUUGGGAUUCAGAUGCAGCUAUAGCUGCAUAUAGCUUUGGAUGC .((((((((........))))))))........((((((((((((((((.(((((....))))))))))..(((((((....))))))).)))))..)))))) (-42.87 = -43.20 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:40 2006