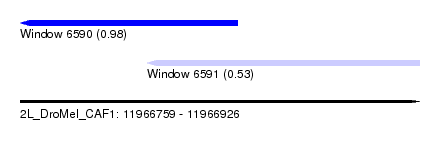
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,966,759 – 11,966,926 |
| Length | 167 |
| Max. P | 0.984190 |

| Location | 11,966,759 – 11,966,850 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 71.71 |
| Mean single sequence MFE | -24.69 |
| Consensus MFE | -16.31 |
| Energy contribution | -17.45 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
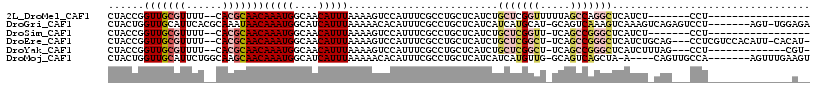
>2L_DroMel_CAF1 11966759 91 - 22407834 CUACCGGUUGCGUUUU--CACGCAACAAAUGGCAACAUUUAAAAGUCCAUUUCGCCUGCUCAUCUGCUCGGUUUUUAGCCAGGCUCAUCU-------CCU----------------- ......(((((((...--.)))))))(((((....))))).............((((((......))..(((.....)))))))......-------...----------------- ( -24.40) >DroGri_CAF1 170260 108 - 1 CUACUGGUUGCAUUCACGCAAAUAACAAAUGGCAUCAUUUAAAAACACAUUUCGCCUGCUCAUCAUCAUGCAU-GCAGUCAAAGUCAAAGUCAGAGUCCU-------AGU-UGGAGA ...((((((((......)))).....(((((....))))).............((.(((..........))).-))..............))))..(((.-------...-.))).. ( -14.60) >DroSim_CAF1 152506 90 - 1 CUACCGGUUGCGUUUU--CACGCAACAAAUGGCAACAUUUAAAAGUCCAUUUCGCCUGCUCAUCUGCUCGGUU-UCAGCCGGGCUCAUCU-------CCU----------------- ......(((((((...--.)))))))(((((....))))).........................(((((((.-...)))))))......-------...----------------- ( -25.60) >DroEre_CAF1 160842 109 - 1 CUACCGGUUGCGUUUU--CACGCAACAAAUGGCAACAUUUAAAAGUCCAUUUCGCCUGCUCAUCUGCUCGGCU-UCAGCCGGGCUCAUCUGCAG---CCUCGUCCACAUU-CACAU- ......(((((((...--.)))))))(((((....))))).............(.((((..((..(((((((.-...)))))))..))..))))---.)...........-.....- ( -33.80) >DroYak_CAF1 157320 97 - 1 CUACCGGUUGCGUUUU--CACGCAACAAAUGGCAACAUUUAAAAGUCCAUUUCGCCUGCUCAUCUGCUCGGCU-UCAGCCGGGCUCAUCUUUAG---CCU-------------CGU- ......(((((((...--.)))))))(((((....))))).................(((.....(((((((.-...)))))))........))---)..-------------...- ( -28.42) >DroMoj_CAF1 178237 104 - 1 CUACUGGUUGCAUUCUGGCAAGCAACAAAUGGCAUCAUUUAAAAACACAUUUCGCCUGCUCAUCAUCAUGUUG-GCAGUCAGCUA-A----CAGUUGCCA-------AGUUUGAAGU ....((.((((......)))).)).(((((((((.(.................(....).........(((((-((.....))))-)----))).)))))-------..)))).... ( -21.30) >consensus CUACCGGUUGCGUUUU__CACGCAACAAAUGGCAACAUUUAAAAGUCCAUUUCGCCUGCUCAUCUGCUCGGUU_UCAGCCAGGCUCAUCU_CAG___CCU______________A__ ......(((((((......)))))))(((((....))))).........................(((((((.....)))))))................................. (-16.31 = -17.45 + 1.14)
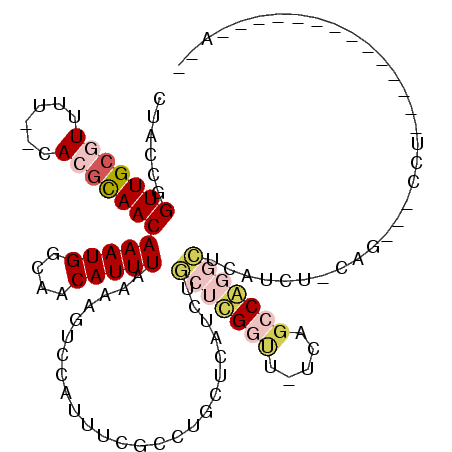
| Location | 11,966,812 – 11,966,926 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 89.41 |
| Mean single sequence MFE | -30.71 |
| Consensus MFE | -21.80 |
| Energy contribution | -23.08 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11966812 114 - 22407834 -AGAGGCAAUCCCGGGAAACGUGGCAAUUCCGAAACCCUCCGCAGCCCAAGAAAUCGCAAUGCUGAACGUUUUUUGGCUACCGGUUGCGUUUUCACGCAACAAAUGGCAACAUUU -...((.....(((((((((((((.....))...........((((....(....).....)))).)))))))))))...)).(((((((....)))))))(((((....))))) ( -30.40) >DroSec_CAF1 186219 114 - 1 -AGCGGCAAUCCCUGGAAACAUGGCAAUUCUCAAACCCUCUGCAGCUAAAGAAAUCGCAAUGCUGAACGUUUUUUGGCUACCGGUUGCGUUUUCACGCAACAAAUGGCAACAUUU -..(((....((.((....)).))...................((((((((((....((....))....)))))))))).)))(((((((....)))))))(((((....))))) ( -31.50) >DroSim_CAF1 152558 114 - 1 -AGCGGCAAUCCCUGGAAACAUGGCAAUUCCGAAACCCUCCGCAGCUAAAGAAAUCGCAAUGCUGAACGUUUUUUGGCUACCGGUUGCGUUUUCACGCAACAAAUGGCAACAUUU -.((((.......(((((.........))))).......))))((((((((((....((....))....))))))))))....(((((((....)))))))(((((....))))) ( -32.74) >DroEre_CAF1 160913 107 - 1 -----CCAAUCCCAGGA-ACAUAGCAAUUGCAAACC-CUCCUCAGCCGAAG-AAUCGCAAUGCUGAACGUUUUUUGGCUACCGGUUGCGUUUUCACGCAACAAAUGGCAACAUUU -----.........((.-.....((....)).....-......((((((((-(((..((....))...))))))))))).)).(((((((....)))))))(((((....))))) ( -29.40) >DroYak_CAF1 157379 114 - 1 CAGUGCCAAUCCGUGGA-ACAUGGCAAUUCCAAACCCCUUCUCAGACCAAGCAAUCGCAAUGCUGAACGUUUUUUGGCUACCGGUUGCGUUUUCACGCAACAAAUGGCAACAUUU ..(((((((..((((((-(........))))..........((((.....((....))....)))))))....))))).))..(((((((....)))))))(((((....))))) ( -29.50) >consensus _AGCGGCAAUCCCUGGAAACAUGGCAAUUCCAAAACCCUCCGCAGCCAAAGAAAUCGCAAUGCUGAACGUUUUUUGGCUACCGGUUGCGUUUUCACGCAACAAAUGGCAACAUUU ..........((.((....)).))...................((((((((((....((....))....))))))))))....(((((((....)))))))(((((....))))) (-21.80 = -23.08 + 1.28)

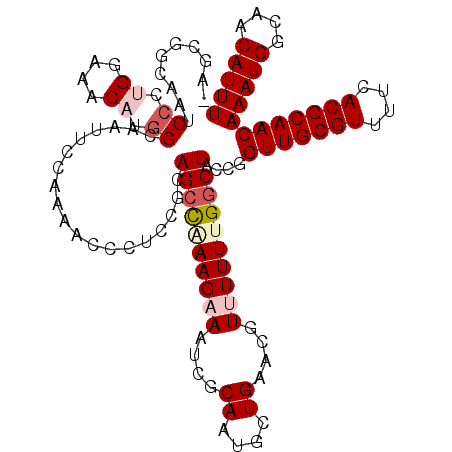
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:30 2006