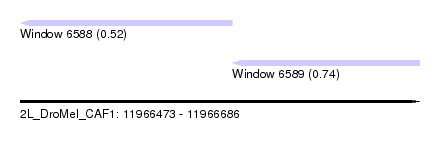
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,966,473 – 11,966,686 |
| Length | 213 |
| Max. P | 0.744314 |

| Location | 11,966,473 – 11,966,586 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 92.47 |
| Mean single sequence MFE | -38.04 |
| Consensus MFE | -33.94 |
| Energy contribution | -33.46 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.89 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11966473 113 - 22407834 GGAGCUGGCUCAAUUGAUGCUGACAGCGAAUGAAAGGCUCUGGCUGAGAGUCUGCAACUGAGACCACGGCCAGAACGGCAAAGCCCUGGGCACAGUGGUUGAAAAACGAUUUG--- ...(((((((((.....(((((...((.........))((((((((.(.((((.......))))).)))))))).)))))......))))).)))).(((....)))......--- ( -37.20) >DroSec_CAF1 185887 113 - 1 GGAGCUGGCUCAAUUGAUGCUGACAGCGAAUGAAAGGCUCUGGCUGAAAGUCUGCAACUGAGACCUCGGCCAGAAUGGCACAGCCCUGGGCACAGUGGUUGAAAAACGAUUUG--- .(((....)))(((((.((((....((.........))(((((((((..((((.......)))).)))))))))..))))((((((((....))).))))).....)))))..--- ( -36.90) >DroSim_CAF1 152223 116 - 1 GGAGCUGGCUCAAUUGAUGCUGACAGCGAAUGAAAGGCUCUGGCUGAGAGUCUGCAACUGAGACCUCGGCCAGAACGGCACAGCCCUGGACACAGUGGUUGAAAAACGAUUUGGUU ......((((.(((((.(((((...((.........))((((((((((.((((.......)))))))))))))).)))))((((((((....))).))))).....))))).)))) ( -44.40) >DroEre_CAF1 160551 113 - 1 GGAGCUGGCUCAAUUGAUGCUGACAGCGAAUGAAAGGCUGUGGCUGAGAGUCUGCAACUGGGACCAUGGCCAGAACAGUGCAGCCCUGGGCACAGUGGUUGAAAAGCGAUUGG--- ...(((((((..((((.(((.....))).......((((((((((.((.........)).)).))))))))....))))..))))...))).((((.(((....))).)))).--- ( -35.80) >DroYak_CAF1 157025 113 - 1 GGAGCUGGCUCAAUUGAUGCUGACAGCGAAUGAAAGGCUGUGGCUGAGCGUCUGCAACUGGGACCAUGGCCAGAACAGUACAGCCCUGGGCACAGUGGUUGGAAAGCGAUUUG--- ...(((((((((.....(((.....))).......(((((((.(((.((....))..((((........))))..)))))))))).))))).)))).(((....)))......--- ( -35.90) >consensus GGAGCUGGCUCAAUUGAUGCUGACAGCGAAUGAAAGGCUCUGGCUGAGAGUCUGCAACUGAGACCACGGCCAGAACGGCACAGCCCUGGGCACAGUGGUUGAAAAACGAUUUG___ .(((....)))(((((.(((((...((.........))((((((((.(.((((.......))))).)))))))).)))))((((((((....))).))))).....)))))..... (-33.94 = -33.46 + -0.48)

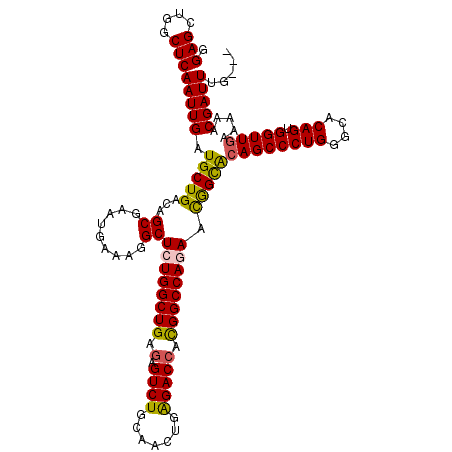
| Location | 11,966,586 – 11,966,686 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 92.84 |
| Mean single sequence MFE | -38.50 |
| Consensus MFE | -35.60 |
| Energy contribution | -35.72 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11966586 100 - 22407834 AACAUAGCACUGGCUCAUAUAUCUUGGGUCUGAAUGGU-----UGGCCGGAGGAGCGGAGAUCUCGACGAUCUUGGGAGCCAUCGGGGCUCGGCUUUUGCGGUGU ....((((...((((((.......))))))......))-----))((((.((((((.((((((.....))))))..(((((.....))))).)))))).)))).. ( -36.70) >DroSec_CAF1 186000 100 - 1 AACAUAGCACUGGCUCAUAUAUCUUGGGCCUGAAUGGU-----UGGCCGGAGGAGCGGAGAUCUCGCCGAUCUUGGGAGCCAUCGGGGCUCGGCUUUUGCGGUGU ....((((...((((((.......))))))......))-----))((((.((((((.((((((.....))))))..(((((.....))))).)))))).)))).. ( -39.40) >DroSim_CAF1 152339 100 - 1 AACAUAGCACUGGCUCAUAUAUCUUGGGCCUGAAUGGU-----UGGCUGGAGGAGCGGAGAUCUCGCCGAUCUUGGGAGCCAUCGGGGCUCGGCUUUUGCGGUGU .((((.(((..((((......((((.((((........-----.)))).))))..(.((((((.....)))))).)(((((.....)))))))))..))).)))) ( -37.40) >DroEre_CAF1 160664 100 - 1 AACAUAGCACUGGCUCAUAUAUCUUGGGCCUGAAUGGC-----UCACCGGAGGAGCGCAGAUCCCGCAGAUCUUGGGAGCCAUCGGGGCUCGGCUUUUGCGGUGU .....(((...((((((.......))))))......))-----)(((((.((((((..(((((.....)))))...(((((.....))))).)))))).))))). ( -39.50) >DroYak_CAF1 157138 105 - 1 AACAUAGCACUGGCUCAUAUAUCUUGGGCCUGAAUGGUUCGGUUCGCCGGAGGAGCGCUGAUCUCCGAGAUCUUGGGAGCCAUCGGGGCUCGGCUUUUGCGGUGU ......((((((((((.....(((.(((((.....)))))((....))))).))))(((((.((((((..((....))....)))))).))))).....)))))) ( -39.50) >consensus AACAUAGCACUGGCUCAUAUAUCUUGGGCCUGAAUGGU_____UGGCCGGAGGAGCGGAGAUCUCGCCGAUCUUGGGAGCCAUCGGGGCUCGGCUUUUGCGGUGU ..(((..((..((((((.......)))))))).))).........((((.((((((.((((((.....))))))..(((((.....))))).)))))).)))).. (-35.60 = -35.72 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:28 2006