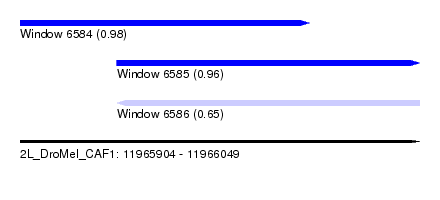
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,965,904 – 11,966,049 |
| Length | 145 |
| Max. P | 0.977340 |

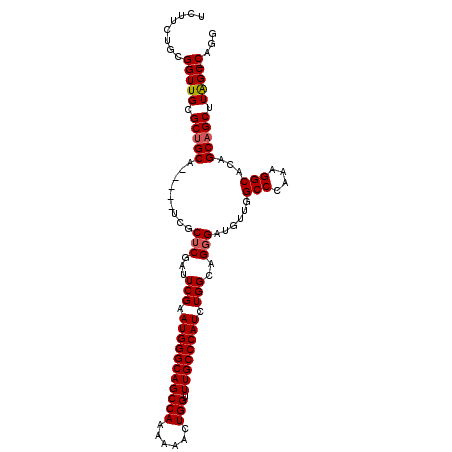
| Location | 11,965,904 – 11,966,009 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 90.73 |
| Mean single sequence MFE | -45.54 |
| Consensus MFE | -34.68 |
| Energy contribution | -34.92 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11965904 105 + 22407834 UCUUUUGCGGUUGCGCUGCA-----UCGCUCGAUUCGAAUGGGCAGCCAAAAAACUGGUUUGCCCAUCUGGCAGGGAUAUUGGCCGAAAGGCACAGCAGCUUAGCCCAGG ....(((.(((((.((((((-----((.((....(((.(((((((((((......))).)))))))).))).)).)))..(((((....))).))))))).)))))))). ( -45.10) >DroSec_CAF1 185292 109 + 1 UCUUCUGCGGUUGCGCUGCAUCGCAUCGCUCGAUUCGAAUGGGCAGCCAAAAAACUGGUUUGCCCAUCUGGCAGGGAUGUUGGCCCAAAGGCACAGCAGCUUAGC-CAGG .....(((((.....)))))(((.(((....))).)))(((((((((((......))).))))))))(((((..((.((((((((....))).))))).))..))-))). ( -47.00) >DroSim_CAF1 151644 109 + 1 UCUUCUGCGGUUGCGCUGCAUCGCAUCGCUCGUUUCGAAUGGGCAGCCAGAAAACUGGUUUGCCCAUCUGGCAGGGAUGUUGGCCCAAAGGCACAGCAGCUUAGC-CAGG ......(((((.(((......)))))))).........((((((((((((....)))).))))))))(((((..((.((((((((....))).))))).))..))-))). ( -48.60) >DroEre_CAF1 160036 104 + 1 UCGGCUGUGGUUGCGCCGCA-----UCGCUCGAUUCGAAUGGGCAGCCAAAAAACUGGUUUGCCCAUCUGGCAGGGAAGUUGGCCCAAAGGCAGAGCAGCUUGGC-CAGG ((((((((((.....)))))-----..)).))).....(((((((((((......))).))))))))(((((..((..(((.(((....)))..)))..))..))-))). ( -43.00) >DroYak_CAF1 156424 104 + 1 ACGGCUGUGGUUGCGCUGCA-----UCGCUCGAUUCGAAUGGGCAGCCAAAAAACUGGUUUGCCCAUCUGGCAGUGAAGUUGGCCCAAAGGCAGAGCAGCUUAGC-CAGG ..(((((.((((((((((((-----((....))).((.(((((((((((......))).)))))))).))))))).......(((....)))...))))))))))-)... ( -44.00) >consensus UCUUCUGCGGUUGCGCUGCA_____UCGCUCGAUUCGAAUGGGCAGCCAAAAAACUGGUUUGCCCAUCUGGCAGGGAUGUUGGCCCAAAGGCACAGCAGCUUAGC_CAGG ........(((((.(((((.........(((...(((.(((((((((((......))).)))))))).)))..)))......(((....)))...))))).)))).)... (-34.68 = -34.92 + 0.24)


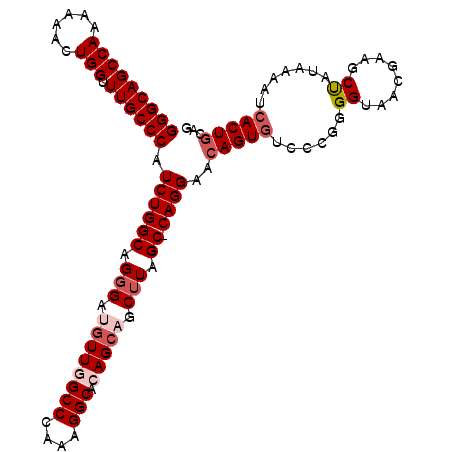
| Location | 11,965,939 – 11,966,049 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 91.77 |
| Mean single sequence MFE | -43.56 |
| Consensus MFE | -31.58 |
| Energy contribution | -33.02 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.61 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960726 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11965939 110 + 22407834 GGGCAGCCAAAAAACUGGUUUGCCCAUCUGGCAGGGAUAUUGGCCGAAAGGCACAGCAGCUUAGCCCAGGAACAGUGUCCCGGGGUAACGAAGCUAUAAAAUCACUGCAG (((((((((......))).)))))).....((((.(((.((((((....))).))).(((((.((((.(((......)))..))))....))))).....))).)))).. ( -44.20) >DroSec_CAF1 185332 109 + 1 GGGCAGCCAAAAAACUGGUUUGCCCAUCUGGCAGGGAUGUUGGCCCAAAGGCACAGCAGCUUAGC-CAGGAACAGUGUCCCGUGGUAACGAAGCUAUAAAAUCACUGCAG (((((((((......))).)))))).((((((..((.((((((((....))).))))).))..))-))))..(((((....(((((......))))).....)))))... ( -44.90) >DroSim_CAF1 151684 109 + 1 GGGCAGCCAGAAAACUGGUUUGCCCAUCUGGCAGGGAUGUUGGCCCAAAGGCACAGCAGCUUAGC-CAGGAACAGUGUCCCGUGGUAACGAAGCUAUAAAAUAACUGCAG ((((((((((....)))).)))))).((((((..((.((((((((....))).))))).))..))-))))..((((.....(((((......)))))......))))... ( -44.80) >DroEre_CAF1 160071 109 + 1 GGGCAGCCAAAAAACUGGUUUGCCCAUCUGGCAGGGAAGUUGGCCCAAAGGCAGAGCAGCUUGGC-CAGGAACAGUGUCCUGGGGAAACGAAGCUAUAAAAUCACUGGAC (((((((((......))).))))))......(((.((.(((.(((....)))..)))(((((..(-(((((......))))))(....).)))))......)).)))... ( -38.90) >DroYak_CAF1 156459 109 + 1 GGGCAGCCAAAAAACUGGUUUGCCCAUCUGGCAGUGAAGUUGGCCCAAAGGCAGAGCAGCUUAGC-CAGGAACAGUGUCCUGGGGAAACGGGGCCAUAAAAUCACUCGGC (((((((((......))).))))))..(((..(((((...((((((..((((......))))..(-(((((......))))))(....).)))))).....)))))))). ( -45.00) >consensus GGGCAGCCAAAAAACUGGUUUGCCCAUCUGGCAGGGAUGUUGGCCCAAAGGCACAGCAGCUUAGC_CAGGAACAGUGUCCCGGGGUAACGAAGCUAUAAAAUCACUGCAG (((((((((......))).)))))).((((((.(((.((((((((....))).))))).))).)).))))..(((((......((........)).......)))))... (-31.58 = -33.02 + 1.44)


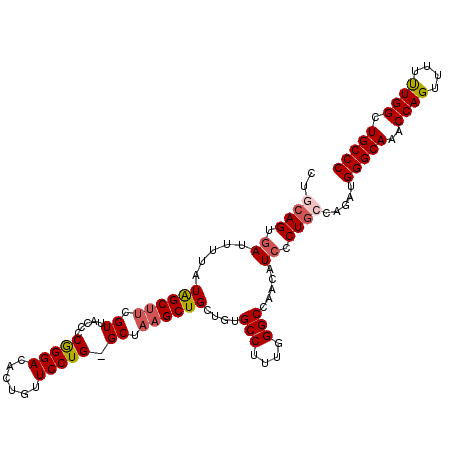
| Location | 11,965,939 – 11,966,049 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 91.77 |
| Mean single sequence MFE | -35.42 |
| Consensus MFE | -27.52 |
| Energy contribution | -28.16 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11965939 110 - 22407834 CUGCAGUGAUUUUAUAGCUUCGUUACCCCGGGACACUGUUCCUGGGCUAAGCUGCUGUGCCUUUCGGCCAAUAUCCCUGCCAGAUGGGCAAACCAGUUUUUUGGCUGCCC ((((((.(((.(((((((..(.(((.(((((((......))))))).))))..)))))(((....))).)).))).))).)))..(((((..((((....)))).))))) ( -39.20) >DroSec_CAF1 185332 109 - 1 CUGCAGUGAUUUUAUAGCUUCGUUACCACGGGACACUGUUCCUG-GCUAAGCUGCUGUGCCUUUGGGCCAACAUCCCUGCCAGAUGGGCAAACCAGUUUUUUGGCUGCCC ((((((.(((...(((((..(.((((((.(((((...)))))))-).))))..)))))(((....)))....))).))).)))..(((((..((((....)))).))))) ( -34.80) >DroSim_CAF1 151684 109 - 1 CUGCAGUUAUUUUAUAGCUUCGUUACCACGGGACACUGUUCCUG-GCUAAGCUGCUGUGCCUUUGGGCCAACAUCCCUGCCAGAUGGGCAAACCAGUUUUCUGGCUGCCC ((((((.......(((((..(.((((((.(((((...)))))))-).))))..)))))(((....)))........))).)))..(((((..((((....)))).))))) ( -34.70) >DroEre_CAF1 160071 109 - 1 GUCCAGUGAUUUUAUAGCUUCGUUUCCCCAGGACACUGUUCCUG-GCCAAGCUGCUCUGCCUUUGGGCCAACUUCCCUGCCAGAUGGGCAAACCAGUUUUUUGGCUGCCC ((((((.((......(((...((((..((((((......)))))-)..)))).)))..(((....))).....)).)))...)))(((((..((((....)))).))))) ( -30.20) >DroYak_CAF1 156459 109 - 1 GCCGAGUGAUUUUAUGGCCCCGUUUCCCCAGGACACUGUUCCUG-GCUAAGCUGCUCUGCCUUUGGGCCAACUUCACUGCCAGAUGGGCAAACCAGUUUUUUGGCUGCCC ((..(((((.....((((((.((((..((((((......)))))-)..)))).((...))....))))))...))))))).....(((((..((((....)))).))))) ( -38.20) >consensus CUGCAGUGAUUUUAUAGCUUCGUUACCCCGGGACACUGUUCCUG_GCUAAGCUGCUGUGCCUUUGGGCCAACAUCCCUGCCAGAUGGGCAAACCAGUUUUUUGGCUGCCC ..((((.((.....((((((.((.....(((((......))))).)).))))))....(((....))).....)).)))).....(((((..((((....)))).))))) (-27.52 = -28.16 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:25 2006