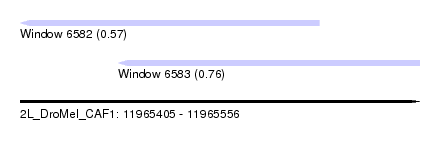
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,965,405 – 11,965,556 |
| Length | 151 |
| Max. P | 0.761998 |

| Location | 11,965,405 – 11,965,518 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.23 |
| Mean single sequence MFE | -21.10 |
| Consensus MFE | -14.61 |
| Energy contribution | -15.05 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
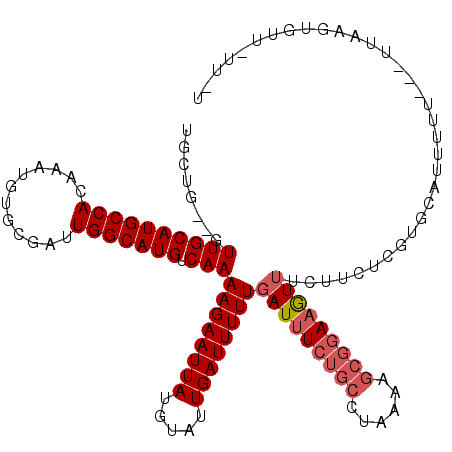
>2L_DroMel_CAF1 11965405 113 - 22407834 AAAGAAUUAUGUAUUGAUUUUUGAUUUCUGCCUAAAAGCGGAAGUUUCUUCUCUUGCGUUUUU---UAAAGUGUUUUUU-UUUGUUAUUUUUUGGUUUUUGUGGCAGUGUCCGCCUC .((((((((.....))))))))(((..(((((.(((((((.(((........))).)))))))---.((((((......-.....))))))...........))))).)))...... ( -20.20) >DroSec_CAF1 184790 103 - 1 AAAGAAUUAUGUAUUGAUUUUUGAUUUCUGCCUAAAAGCGGAAGUUUCUUUUCGUGCAUUUUU---UUAAGUGUUAUUU-----------UUUUUUUUUUGUGUCAGUGCCCGCCUC ..........((((((((....(((((((((......))))))))).........((((((..---..)))))).....-----------............))))))))....... ( -18.80) >DroSim_CAF1 151157 99 - 1 AAAGAAUUAUGUAUUGAUUUUUGAUUUCUGCCUAAAAGCGGAAGUUUCUUCUCAUACAUUUUU---UUAAGUGUU---------------UUUCUUUUUUGUGGCAGUGCCCGCCUC ..((((..((((((.((.....(((((((((......))))))))).....))))))))..))---)).......---------------............(((.......))).. ( -20.40) >DroEre_CAF1 159572 106 - 1 AAAGAAUUAUGUAUUGAUUUUUGAUUUCUGCCUAAAAGCGGAAGUUUCUUCUCCUGCAUUUUUUUUUGUAGUGCUGUUUUCUUGCUG-----------CUGCUGCAGUGCCCGCCCC ((((((..(((((..((.....(((((((((......)))))))))...))...)))))..))))))...(..((((......((..-----------..)).))))..)....... ( -25.00) >consensus AAAGAAUUAUGUAUUGAUUUUUGAUUUCUGCCUAAAAGCGGAAGUUUCUUCUCAUGCAUUUUU___UUAAGUGUU_UUU___________UUU_UUUUUUGUGGCAGUGCCCGCCUC .((((((((.....))))))))(((((((((......))))))))).........(((((((.....)))))))..........................((((......))))... (-14.61 = -15.05 + 0.44)

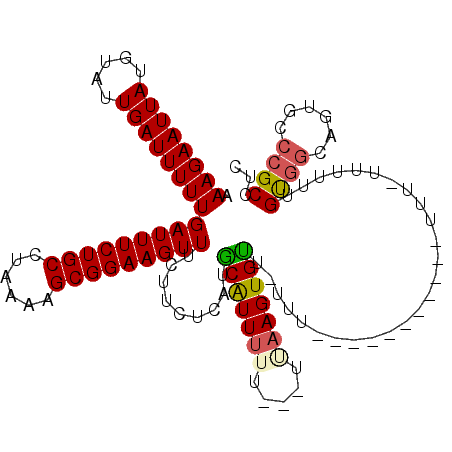
| Location | 11,965,442 – 11,965,556 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.88 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -19.56 |
| Energy contribution | -20.42 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11965442 114 - 22407834 UGCUG--GUUGCAUGCCACAAAUGUGCGAUUGGCAUGUCAAAAGAAUUAUGUAUUGAUUUUUGAUUUCUGCCUAAAAGCGGAAGUUUCUUCUCUUGCGUUUUU---UAAAGUGUUUUU-U (((..--(((((((.........)))))))..)))........(((..(((((..((.....(((((((((......)))))))))...))...)))))..))---)...........-. ( -29.20) >DroVir_CAF1 168773 95 - 1 UGCUGCUGUUGCAUGCCACAAAUGUGCGAUUGGCAUGUCAAAAGAAUUAUGUAUUGAUUUUUGAUUUG--G-C-AUCGUGGAUAUAUUUGG-CGA-CGAUC------------------- ...(((....)))(((((...((.((((((.(.((.(((((..((((((.....))))))))))).))--.-)-)))))).)).....)))-)).-.....------------------- ( -22.70) >DroSec_CAF1 184817 114 - 1 UGCUG--GUUGCAUGCCACAAAUGUGCGAUUGGCAUGUCAAAAGAAUUAUGUAUUGAUUUUUGAUUUCUGCCUAAAAGCGGAAGUUUCUUUUCGUGCAUUUUU---UUAAGUGUUAUU-U (((..--(((((((.........)))))))..))).......((((..((((((.((.....(((((((((......))))))))).....))))))))..))---))..........-. ( -31.60) >DroSim_CAF1 151184 110 - 1 UGCUG--GUUGCAUGCCACAAAUGUGCGAUUGGCAUGUCAAAAGAAUUAUGUAUUGAUUUUUGAUUUCUGCCUAAAAGCGGAAGUUUCUUCUCAUACAUUUUU---UUAAGUGUU----- (((..--(((((((.........)))))))..))).......((((..((((((.((.....(((((((((......))))))))).....))))))))..))---)).......----- ( -31.30) >DroEre_CAF1 159599 117 - 1 UGCUG--GUUGCAUGCCACAAAUGUGCGAUUGGCAUGUCAAAAGAAUUAUGUAUUGAUUUUUGAUUUCUGCCUAAAAGCGGAAGUUUCUUCUCCUGCAUUUUUUUUUGUAGUGCUGUU-U (((..--(((((((.........)))))))..)))...((((((((..(((((..((.....(((((((((......)))))))))...))...))))).))))))))..........-. ( -32.60) >DroYak_CAF1 155948 117 - 1 UGCUG--GUUGCAUGCCACAAAUGUGCGAUUGGCAUGUCAAAAGAAUUAUGUAUUGAUUUUUGAUUUCUGCCUUAAAGCGGAAGUUUCUUCUCUU-CUUAUUUUUUUUUAGUGUUGAUUU (((..--(((((((.........)))))))..))).(((((((((((((.....))))))))(((((((((......))))))))).........-.................))))).. ( -28.90) >consensus UGCUG__GUUGCAUGCCACAAAUGUGCGAUUGGCAUGUCAAAAGAAUUAUGUAUUGAUUUUUGAUUUCUGCCUAAAAGCGGAAGUUUCUUCUCGUGCAUUUUU___UUAAGUGUU_UU_U ........((((((((((............))))))).)))((((((((.....))))))))(((((((((......))))))))).................................. (-19.56 = -20.42 + 0.86)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:23 2006