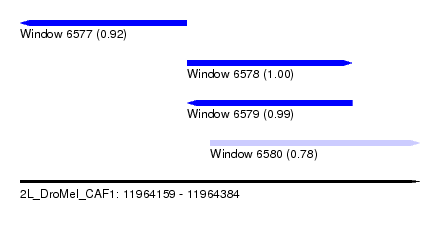
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,964,159 – 11,964,384 |
| Length | 225 |
| Max. P | 0.997886 |

| Location | 11,964,159 – 11,964,253 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 92.41 |
| Mean single sequence MFE | -23.49 |
| Consensus MFE | -19.44 |
| Energy contribution | -19.92 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.923078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11964159 94 - 22407834 GAUCUCGUAGAUAAAAGUGGCUUAAACUAUGUAAGCCAUCAACGUUUUAUGAGAGUGGUAUAAACAA--CAGAUACGACUUAGAACGCUCUGGUGA ..(((((((((.....((((((((.......))))))))......)))))))))((.((((......--...)))).))......(((....))). ( -23.70) >DroSec_CAF1 183584 94 - 1 GAUCUCGUAGAUAAAAGUGGCUUAAACUAUGUAAGCCAUCAACGUUUUAUGAGAGUCUUAUAAAGCA--CAGAUACGACUUAGAACGCUCUAGUGA ....(((((.......((((((((.......))))))))....((((((((((...)))))))))).--....))))).......(((....))). ( -22.80) >DroSim_CAF1 149961 94 - 1 GAUCUCGUAGAUAAAAGUGGCUUAAACUAUGUAAGCCAUCAACGUUUUAUGAGAGUGGUAUAAAGCA--CAGAUACGACUUAGAACGCUCUGGUGA ..(((((((((.....((((((((.......))))))))......)))))))))((.((((......--...)))).))......(((....))). ( -23.70) >DroEre_CAF1 158432 94 - 1 GAUCUCGCAGAUAAAAGUGGCUUAAACUAUGUAAGCCAUCAGCGUUUUAUGAGAGUGGCAUAAUGAA--CAGAUACGACUUAGAACGCUCUGGUGA ....(((((((.....((((((((.......))))))))..((((((((.....(((..........--....)))....))))))))))).)))) ( -22.04) >DroYak_CAF1 154691 96 - 1 GAUCUCGUAGAUAAAGAUGGCUUAAACUAUGUAAGCCAUCAGCGUUUUAUGAGUGUGGUAUAAAGAGCGCAGAUACGACUUAGAAUGCUGUGGUGA .(((.(.........(((((((((.......)))))))))(((((((((.....((.((((...........)))).)).)))))))))).))).. ( -25.20) >consensus GAUCUCGUAGAUAAAAGUGGCUUAAACUAUGUAAGCCAUCAACGUUUUAUGAGAGUGGUAUAAAGAA__CAGAUACGACUUAGAACGCUCUGGUGA ..(((((((((.....((((((((.......))))))))......)))))))))((.((((...........)))).))......(((....))). (-19.44 = -19.92 + 0.48)


| Location | 11,964,253 – 11,964,346 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 84.30 |
| Mean single sequence MFE | -28.12 |
| Consensus MFE | -22.48 |
| Energy contribution | -23.10 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.57 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.997886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11964253 93 + 22407834 UGCCAUUCCCUGGCCCUCUCCUCGGGCUCAAUAUACAUAUAUAUAUACAUAGAUACAUAGCACAAUAGAUCGAUCUGAUGAGAGGGCCAGCUA .........(((((((((((.(((((.((.(((((......))))).....(((...((......)).)))))))))).)))))))))))... ( -31.40) >DroSec_CAF1 183678 89 + 1 UGCCAUUCUCUGGCCCUCUCCUCGGGCUCAAGAUACAUA----UAUACAUAGAUACAUAGUACAAUAGAUCGAUCUGAUGAGAGGGCCAGCUA .........(((((((((((.(((((.((..(((...((----(.(((.((......)))))..))).)))))))))).)))))))))))... ( -29.70) >DroSim_CAF1 150055 89 + 1 UGCCAUUCUCUGGCCCUCUCCUCGGGCUCAAGAUACAUA----UAUACAUAGAUACAUAGUACAAUAGAUCGAUCUGUUGAGAGGGCCGGCUA .........(((((((((((..((((.((..(((...((----(.(((.((......)))))..))).)))))))))..)))))))))))... ( -27.50) >DroYak_CAF1 154787 89 + 1 UGCCAUUCUCUGGCCCUCUCCUCGGGCUUGAAA-AUGUAUAUACCUCCAUACAUAUUUAGCACAAUAGAUCGAUCU---GAGAGGCCCAGUCU .........((((.((((((.(((((((.(((.-((((((........)))))).)))))).(....).))))...---)))))).))))... ( -23.90) >consensus UGCCAUUCUCUGGCCCUCUCCUCGGGCUCAAGAUACAUA____UAUACAUAGAUACAUAGCACAAUAGAUCGAUCUGAUGAGAGGGCCAGCUA .........(((((((((((.(((((.((..........................................))))))).)))))))))))... (-22.48 = -23.10 + 0.62)


| Location | 11,964,253 – 11,964,346 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 84.30 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -21.92 |
| Energy contribution | -23.05 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.41 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11964253 93 - 22407834 UAGCUGGCCCUCUCAUCAGAUCGAUCUAUUGUGCUAUGUAUCUAUGUAUAUAUAUAUGUAUAUUGAGCCCGAGGAGAGGGCCAGGGAAUGGCA ...(((((((((((......(((..((..((((((((((((........))))))).)))))...))..))).)))))))))))......... ( -34.60) >DroSec_CAF1 183678 89 - 1 UAGCUGGCCCUCUCAUCAGAUCGAUCUAUUGUACUAUGUAUCUAUGUAUA----UAUGUAUCUUGAGCCCGAGGAGAGGGCCAGAGAAUGGCA ...(((((((((((......(((..((...(((((((((((....)))))----)).))))....))..))).)))))))))))......... ( -32.60) >DroSim_CAF1 150055 89 - 1 UAGCCGGCCCUCUCAACAGAUCGAUCUAUUGUACUAUGUAUCUAUGUAUA----UAUGUAUCUUGAGCCCGAGGAGAGGGCCAGAGAAUGGCA ..((((((((((((......(((..((...(((((((((((....)))))----)).))))....))..))).))))))))).......))). ( -31.11) >DroYak_CAF1 154787 89 - 1 AGACUGGGCCUCUC---AGAUCGAUCUAUUGUGCUAAAUAUGUAUGGAGGUAUAUACAU-UUUCAAGCCCGAGGAGAGGGCCAGAGAAUGGCA ...((((.((((((---(((....))).(((.(((....(((((((......)))))))-.....))).))).)))))).))))......... ( -29.30) >consensus UAGCUGGCCCUCUCAUCAGAUCGAUCUAUUGUACUAUGUAUCUAUGUAUA____UAUGUAUCUUGAGCCCGAGGAGAGGGCCAGAGAAUGGCA ...(((((((((((...(((....))).(((.(((.......((((((......)))))).....))).))).)))))))))))......... (-21.92 = -23.05 + 1.13)

| Location | 11,964,266 – 11,964,384 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.06 |
| Mean single sequence MFE | -28.75 |
| Consensus MFE | -19.99 |
| Energy contribution | -20.93 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.776909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11964266 118 + 22407834 CCCUCUCCUCGGGCUCAAUAUACAUAUAUAUAUACAUAGAUACAUAGCACAAUAGAUCGAUCUGAUGAGAGGGCCAGCUAGGAACAUUCUCUUU--UUUCAAGAGCUUAGUUUUAGCGCA (((((((.(((((.((.(((((......))))).....(((...((......)).)))))))))).)))))))...(((((((.....(((((.--....))))).....)))))))... ( -31.20) >DroSec_CAF1 183691 114 + 1 CCCUCUCCUCGGGCUCAAGAUACAUA----UAUACAUAGAUACAUAGUACAAUAGAUCGAUCUGAUGAGAGGGCCAGCUAGGAACAUUCUCUUU--UUUCAAGAGCUUAGUUUUAGCACA (((((((.(((((.((..(((...((----(.(((.((......)))))..))).)))))))))).)))))))...(((((((.....(((((.--....))))).....)))))))... ( -29.70) >DroSim_CAF1 150068 114 + 1 CCCUCUCCUCGGGCUCAAGAUACAUA----UAUACAUAGAUACAUAGUACAAUAGAUCGAUCUGUUGAGAGGGCCGGCUAGGAACAUUCUCUUU--UUUCAAGAGCUUAGUUUUAGCGCA (((((((..((((.((..(((...((----(.(((.((......)))))..))).)))))))))..)))))))...(((((((.....(((((.--....))))).....)))))))... ( -28.00) >DroYak_CAF1 154800 116 + 1 CCCUCUCCUCGGGCUUGAAA-AUGUAUAUACCUCCAUACAUAUUUAGCACAAUAGAUCGAUCU---GAGAGGCCCAGUCUGGAACAUUCUCUUUUUUUUCAAGAGCUUAGUUUUAGUGCU .((((((.(((((((.(((.-((((((........)))))).)))))).(....).))))...---))))))...(((((((((((..(((((.......)))))..).))))))).))) ( -26.10) >consensus CCCUCUCCUCGGGCUCAAGAUACAUA____UAUACAUAGAUACAUAGCACAAUAGAUCGAUCUGAUGAGAGGGCCAGCUAGGAACAUUCUCUUU__UUUCAAGAGCUUAGUUUUAGCGCA (((((((.(((((.((..........................................))))))).)))))))...(((((((.....(((((.......))))).....)))))))... (-19.99 = -20.93 + 0.94)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:20 2006