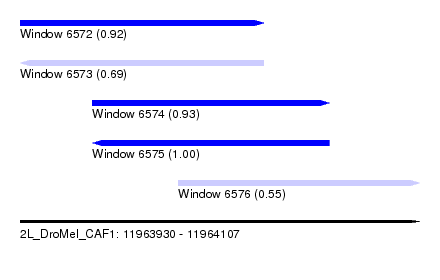
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,963,930 – 11,964,107 |
| Length | 177 |
| Max. P | 0.998931 |

| Location | 11,963,930 – 11,964,038 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.47 |
| Mean single sequence MFE | -31.98 |
| Consensus MFE | -19.14 |
| Energy contribution | -19.14 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11963930 108 + 22407834 A--------UCCCGCUUUUGGGCCAUUCUGAACUUCAUUAGCGACUGCGAAUCAACAA--AGCUGAUGAGCAGCUUAGGGCCAACAGA--GCGACUUGUGGCGAGGAAACAGAAAUGGUA .--------(((((((....((((...(((....((((((((...((........)).--.)))))))).))).....)))).((((.--.....)))))))).)))..((....))... ( -30.70) >DroSec_CAF1 183369 100 + 1 A--------UCCCGCUUUUGGGCCAUUCUGAACUUCAUUAGCGACUGCGAAUCAACAA--AGCUGAUGAGCAGCUUAGGGCCAACAGA--GCGCCUAGUGGUGAGGAA-CA-------UA .--------(((((((.((((((..(((((...(((((((((...((........)).--.)))))))))..((.....))...))))--).)))))).)))).))).-..-------.. ( -32.10) >DroSim_CAF1 149755 86 + 1 A--------UCCCGCUUUUGGGCCAUUCUGAACUUCAUUAGCGACUGCGAAUCAACAA--AGCUGAUGAGCAGCUUAGGGCCAACAGA----------------GGAA-AA-------UA .--------..((.((.((((.(((..(((....((((((((...((........)).--.)))))))).)))..).)).)))).)).----------------))..-..-------.. ( -24.80) >DroEre_CAF1 158237 84 + 1 A--------UCCCGCUUGUGGGCCAUUCUGAACUUCAUUAGCGACUGCGAAUCAACAA--AGCUGAUGAGCAGCUUAGGGCCCACAGA--GCGCCU------------------------ .--------...((((((((((((...(((....((((((((...((........)).--.)))))))).))).....)))))))).)--)))...------------------------ ( -33.60) >DroYak_CAF1 154440 120 + 1 AUCGCAUCAUCCAGCUUUUGGGCCAUUCUGAACUUCAUUAGCGGCUGCGAAUCAACAAAUAGCUGAUGAGCAGCUUAGGGCCCACAGAGAGCGCCUUGAGGUGAUGAAAUACAAAUGGUA (((((.(((....(((((((((((...((((......)))).((((((..((((.(.....).))))..))))))...))))))...)))))....))).)))))............... ( -38.70) >consensus A________UCCCGCUUUUGGGCCAUUCUGAACUUCAUUAGCGACUGCGAAUCAACAA__AGCUGAUGAGCAGCUUAGGGCCAACAGA__GCGCCU_G_GG_GAGGAA_CA_______UA ....................((((...(((....((((((((...((........))....)))))))).))).....))))...................................... (-19.14 = -19.14 + 0.00)

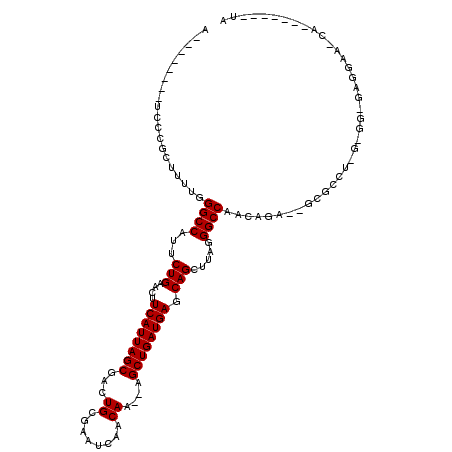

| Location | 11,963,930 – 11,964,038 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.47 |
| Mean single sequence MFE | -30.14 |
| Consensus MFE | -21.94 |
| Energy contribution | -22.38 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.693886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11963930 108 - 22407834 UACCAUUUCUGUUUCCUCGCCACAAGUCGC--UCUGUUGGCCCUAAGCUGCUCAUCAGCU--UUGUUGAUUCGCAGUCGCUAAUGAAGUUCAGAAUGGCCCAAAAGCGGGA--------U ......((((((((....((((......((--(..((((((.....(((((..((((((.--..))))))..))))).))))))..)))......))))....))))))))--------. ( -31.20) >DroSec_CAF1 183369 100 - 1 UA-------UG-UUCCUCACCACUAGGCGC--UCUGUUGGCCCUAAGCUGCUCAUCAGCU--UUGUUGAUUCGCAGUCGCUAAUGAAGUUCAGAAUGGCCCAAAAGCGGGA--------U ..-------..-.......(((.(..(.((--(..((((((.....(((((..((((((.--..))))))..))))).))))))..))).)..).)))(((......))).--------. ( -26.40) >DroSim_CAF1 149755 86 - 1 UA-------UU-UUCC----------------UCUGUUGGCCCUAAGCUGCUCAUCAGCU--UUGUUGAUUCGCAGUCGCUAAUGAAGUUCAGAAUGGCCCAAAAGCGGGA--------U ..-------..-((((----------------.((.((((......(((((..((((((.--..))))))..))))).((((.((.....))...)))))))).)).))))--------. ( -23.30) >DroEre_CAF1 158237 84 - 1 ------------------------AGGCGC--UCUGUGGGCCCUAAGCUGCUCAUCAGCU--UUGUUGAUUCGCAGUCGCUAAUGAAGUUCAGAAUGGCCCACAAGCGGGA--------U ------------------------...(((--(.((((((((....(((((..((((((.--..))))))..))))).(((.....))).......))))))))))))...--------. ( -32.70) >DroYak_CAF1 154440 120 - 1 UACCAUUUGUAUUUCAUCACCUCAAGGCGCUCUCUGUGGGCCCUAAGCUGCUCAUCAGCUAUUUGUUGAUUCGCAGCCGCUAAUGAAGUUCAGAAUGGCCCAAAAGCUGGAUGAUGCGAU ......(((((..(((((.((....)).(((.....((((((....(((((..((((((.....))))))..))))).(((.....))).......))))))..)))..)))))))))). ( -37.10) >consensus UA_______UG_UUCCUC_CC_C_AGGCGC__UCUGUUGGCCCUAAGCUGCUCAUCAGCU__UUGUUGAUUCGCAGUCGCUAAUGAAGUUCAGAAUGGCCCAAAAGCGGGA________U ................................((((((((((....(((((..((((((.....))))))..))))).(((.....))).......))))....)))))).......... (-21.94 = -22.38 + 0.44)

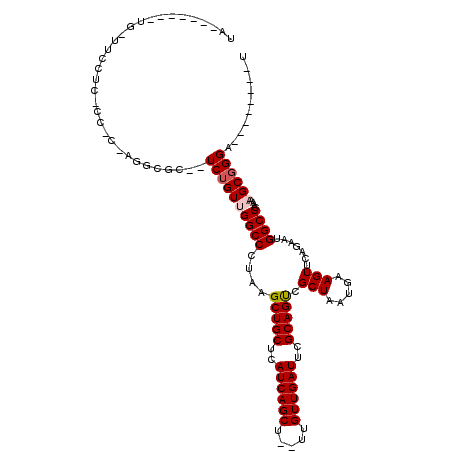
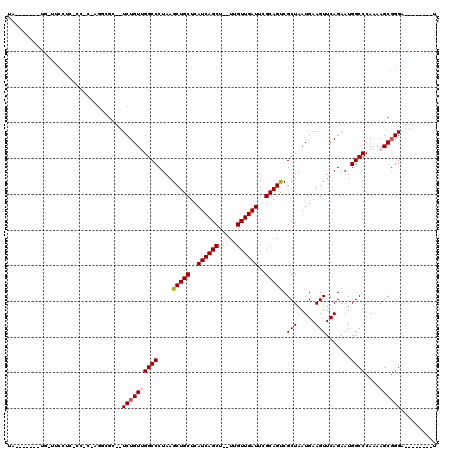
| Location | 11,963,962 – 11,964,067 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 74.85 |
| Mean single sequence MFE | -28.43 |
| Consensus MFE | -17.15 |
| Energy contribution | -19.40 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.927865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11963962 105 + 22407834 GCGACUGCGAAUCAACAA--AGCUGAUGAGCAGCUUAGGGCCAACAGA--GCGACUUGUGGCGAGGAAACAGAAAUGGUAUAUGGCACAAUAC--CAUACCA---UAUACCAUU .((.(..(((.((..(.(--(((((.....)))))).).(((....).--)))).)))..))).(....)...((((((((((((........--....)))---))))))))) ( -31.70) >DroSec_CAF1 183401 86 + 1 GCGACUGCGAAUCAACAA--AGCUGAUGAGCAGCUUAGGGCCAACAGA--GCGCCUAGUGGUGAGGAA-CA-------UAUA-------------UAUACCA---UAUACCAUU ..(.((((..((((....--...))))..))))).(((((((....).--)).))))((((((.((..-..-------....-------------....)).---..)))))). ( -18.60) >DroYak_CAF1 154480 111 + 1 GCGGCUGCGAAUCAACAAAUAGCUGAUGAGCAGCUUAGGGCCCACAGAGAGCGCCUUGAGGUGAUGAAAUACAAAUGGUAUAUGGUGUG---CCAUAUACUAUACUAAACCAUU ...(((((..((((.(.....).))))..)))))(((((((.(.......).)))))))(((..((.....)).(((((((((((....---))))))))))).....)))... ( -35.00) >consensus GCGACUGCGAAUCAACAA__AGCUGAUGAGCAGCUUAGGGCCAACAGA__GCGCCUUGUGGUGAGGAAACA_AAAUGGUAUAUGG_______C__UAUACCA___UAUACCAUU ..(.((((..((((.(.....).))))..)))))...((((((..((.......))..)))).............((((((((((.......)))))))))).......))... (-17.15 = -19.40 + 2.25)

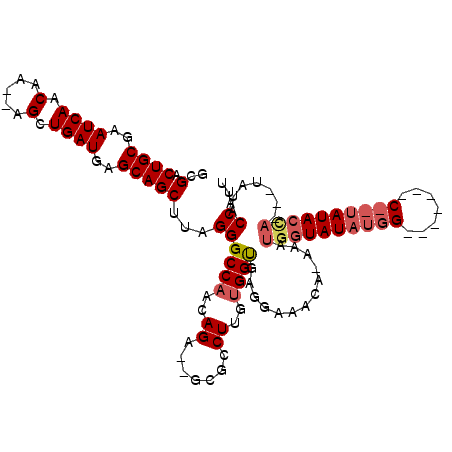

| Location | 11,963,962 – 11,964,067 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 74.85 |
| Mean single sequence MFE | -31.43 |
| Consensus MFE | -18.22 |
| Energy contribution | -19.00 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.58 |
| SVM decision value | 3.29 |
| SVM RNA-class probability | 0.998931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11963962 105 - 22407834 AAUGGUAUA---UGGUAUG--GUAUUGUGCCAUAUACCAUUUCUGUUUCCUCGCCACAAGUCGC--UCUGUUGGCCCUAAGCUGCUCAUCAGCU--UUGUUGAUUCGCAGUCGC (((((((((---((((((.--.....)))))))))))))))...........((((..((....--.))..)))).....(((((..((((((.--..))))))..)))))... ( -36.00) >DroSec_CAF1 183401 86 - 1 AAUGGUAUA---UGGUAUA-------------UAUA-------UG-UUCCUCACCACUAGGCGC--UCUGUUGGCCCUAAGCUGCUCAUCAGCU--UUGUUGAUUCGCAGUCGC ..((((...---.((.(((-------------....-------))-).))..)))).((((.((--(.....))))))).(((((..((((((.--..))))))..)))))... ( -21.60) >DroYak_CAF1 154480 111 - 1 AAUGGUUUAGUAUAGUAUAUGG---CACACCAUAUACCAUUUGUAUUUCAUCACCUCAAGGCGCUCUCUGUGGGCCCUAAGCUGCUCAUCAGCUAUUUGUUGAUUCGCAGCCGC ..((((..((((((((((((((---....))))))))....))))))..)))).....(((.((((.....)))))))..(((((..((((((.....))))))..)))))... ( -36.70) >consensus AAUGGUAUA___UGGUAUA__G_______CCAUAUACCAUUU_UGUUUCCUCACCACAAGGCGC__UCUGUUGGCCCUAAGCUGCUCAUCAGCU__UUGUUGAUUCGCAGUCGC ...((.........(((((((((.....)))))))))................(((..((.......))..))).))...(((((..((((((.....))))))..)))))... (-18.22 = -19.00 + 0.78)



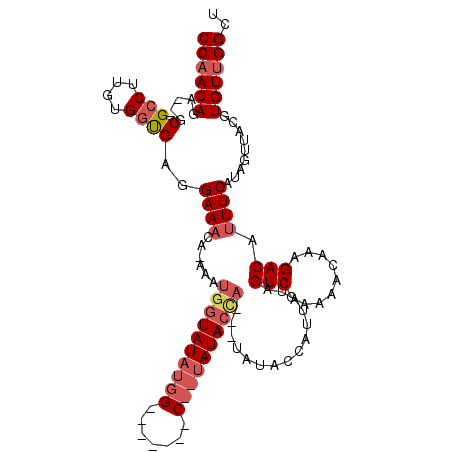
| Location | 11,964,000 – 11,964,107 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 76.20 |
| Mean single sequence MFE | -25.33 |
| Consensus MFE | -13.81 |
| Energy contribution | -16.73 |
| Covariance contribution | 2.92 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.553681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11964000 107 + 22407834 CCAACAGA--GCGACUUGUGGCGAGGAAACAGAAAUGGUAUAUGGCACAAUAC--CAUACCA---UAUACCAUUAUAGUCGAAAACAAAGACAUUUCAUAGUUACGUGUUGGCU ((((((..--(..(((.((((...(....)...((((((((((((........--....)))---)))))))))...(((.........)))...)))))))..).)))))).. ( -30.70) >DroSec_CAF1 183439 88 + 1 CCAACAGA--GCGCCUAGUGGUGAGGAA-CA-------UAUA-------------UAUACCA---UAUACCAUUAUAGUCGAAAACAAAGACAUUUCAUAGUUACGUGUUGGCU ((((((.(--((...((((((((.((..-..-------....-------------....)).---..))))))))..(((.........)))........)))...)))))).. ( -17.10) >DroYak_CAF1 154520 111 + 1 CCCACAGAGAGCGCCUUGAGGUGAUGAAAUACAAAUGGUAUAUGGUGUG---CCAUAUACUAUACUAAACCAUUAUGGUCGAAAACAAAGACAUUUCAUAGUUACGUGUUGGCU ((.(((...((((((....))).(((((((....(((((((((((....---)))))))))))..............(((.........)))))))))).)))...))).)).. ( -28.20) >consensus CCAACAGA__GCGCCUUGUGGUGAGGAAACA_AAAUGGUAUAUGG_______C__UAUACCA___UAUACCAUUAUAGUCGAAAACAAAGACAUUUCAUAGUUACGUGUUGGCU ((((((.....((((....))))..((((......((((((((((.......))))))))))...............(((.........))).)))).........)))))).. (-13.81 = -16.73 + 2.92)

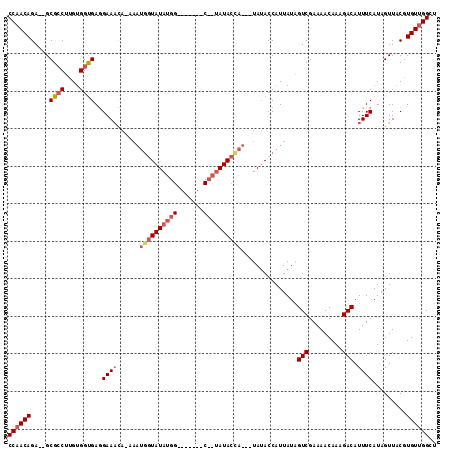
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:16 2006