
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
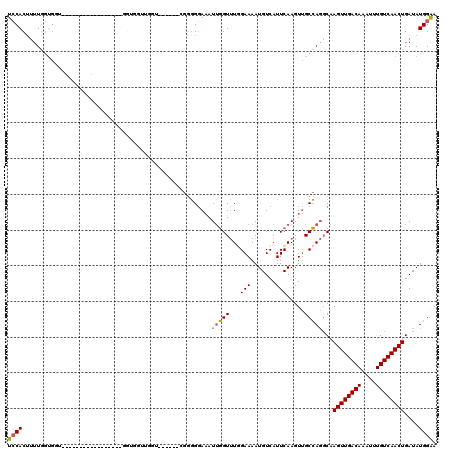
| Location | 11,960,912 – 11,961,012 |
| Length | 100 |
| Max. P | 0.992205 |

| Location | 11,960,912 – 11,961,012 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.34 |
| Mean single sequence MFE | -25.05 |
| Consensus MFE | -15.20 |
| Energy contribution | -15.28 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
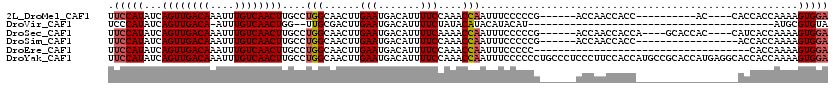
>2L_DroMel_CAF1 11960912 100 + 22407834 UCCACUUUUGGUGGUG----GU----------GGUGGUUGGU------CGGGGGAAAUUGGUUUGGAAAAUGUCAUUCAAGUUGCCAGGCAAGUUGACAAAUUUGUCAACUGAUAUGGAA .(((((......))))----).----------........((------(..((.((.((((..(((......))).)))).)).)).))).((((((((....))))))))......... ( -24.10) >DroVir_CAF1 165256 76 + 1 UACACGCAU-----------------------------------------AUGUAUGUAUGUAUAGAAAAUGUCAUUCAAGUCGCAA--CCAGUUGACAAAU-UGUCAACUGAUAUGGGA ((((..(((-----------------------------------------....)))..))))..................((.((.--.((((((((....-.))))))))...)).)) ( -18.20) >DroSec_CAF1 180322 106 + 1 UCCACUUUUGGUGAUG----GUGGUGC----UGGUGGUUGGU------CGGGGGAAAUUGGUUUUGAAAAUGUCAUUCAAGUUGCCAGGCAAGUUGACAAAUUUGUCAACUGAUAUGGAA ((((..(((((..((.----.(((...----((..(.....(------(((((........))))))...)..)).))).))..)))))..((((((((....))))))))....)))). ( -25.10) >DroSim_CAF1 146706 97 + 1 UCCACUUUUGGUGGU-----------------GGUGGUUGGU------CGGGGGAAAUUGGUUUGGAAAAUGUCAUUCAAGUUGCCAGGCAAGUUGACAAAUUUGUCAACUGAUAUGGAA .(((((......)))-----------------))......((------(..((.((.((((..(((......))).)))).)).)).))).((((((((....))))))))......... ( -24.10) >DroEre_CAF1 155238 84 + 1 UCCACUUUUGGUG------------------------------------GGGGGAAAUUGGUUUGGAAAAUGUCAUUCAAGUUGCCAGGCAAGUUGACAAAUUUGUCAACUGAUAUGGAA (((((.....)))------------------------------------)).......(..((((((........))))))..)(((....((((((((....))))))))....))).. ( -22.00) >DroYak_CAF1 151109 120 + 1 UCCACUUUUGGUGGUGCCUCAUGGUGCGGCAUGGUGGAAGGGAGGGCAGGGGGGAAAUUGGUUUGGAAAAUGUCAUUCAAGUUGCCAGGCAAGUUGACAAAUUUGUCAACUGAUAUGGAA ((((((....((.(..((....))..).))..)))))).......((..((.((...((((..(((......))).)))).)).))..)).((((((((....))))))))......... ( -36.80) >consensus UCCACUUUUGGUGGU_________________GGUGGUUGGU______CGGGGGAAAUUGGUUUGGAAAAUGUCAUUCAAGUUGCCAGGCAAGUUGACAAAUUUGUCAACUGAUAUGGAA ((((.....................................................(((((...(((.......))).....)))))...((((((((....))))))))....)))). (-15.20 = -15.28 + 0.08)


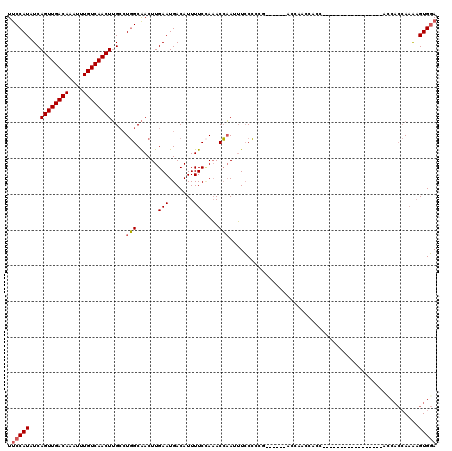
| Location | 11,960,912 – 11,961,012 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.34 |
| Mean single sequence MFE | -18.22 |
| Consensus MFE | -12.46 |
| Energy contribution | -12.68 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11960912 100 - 22407834 UUCCAUAUCAGUUGACAAAUUUGUCAACUUGCCUGGCAACUUGAAUGACAUUUUCCAAACCAAUUUCCCCCG------ACCAACCACC----------AC----CACCACCAAAAGUGGA ..(((....((((((((....))))))))....)))....................................------..........----------..----..((((.....)))). ( -15.50) >DroVir_CAF1 165256 76 - 1 UCCCAUAUCAGUUGACA-AUUUGUCAACUGG--UUGCGACUUGAAUGACAUUUUCUAUACAUACAUACAU-----------------------------------------AUGCGUGUA ((.((.((((((((((.-....)))))))))--))).))..................((((..(((....-----------------------------------------)))..)))) ( -18.20) >DroSec_CAF1 180322 106 - 1 UUCCAUAUCAGUUGACAAAUUUGUCAACUUGCCUGGCAACUUGAAUGACAUUUUCAAAACCAAUUUCCCCCG------ACCAACCACCA----GCACCAC----CAUCACCAAAAGUGGA .(((((...((((((((....))))))))(((.(((....(((((.......)))))..............(------......).)))----)))....----...........))))) ( -19.10) >DroSim_CAF1 146706 97 - 1 UUCCAUAUCAGUUGACAAAUUUGUCAACUUGCCUGGCAACUUGAAUGACAUUUUCCAAACCAAUUUCCCCCG------ACCAACCACC-----------------ACCACCAAAAGUGGA ..(((....((((((((....))))))))....)))....................................------..........-----------------.((((.....)))). ( -15.50) >DroEre_CAF1 155238 84 - 1 UUCCAUAUCAGUUGACAAAUUUGUCAACUUGCCUGGCAACUUGAAUGACAUUUUCCAAACCAAUUUCCCCC------------------------------------CACCAAAAGUGGA ..(((....((((((((....))))))))....)))..................................(------------------------------------(((.....)))). ( -15.30) >DroYak_CAF1 151109 120 - 1 UUCCAUAUCAGUUGACAAAUUUGUCAACUUGCCUGGCAACUUGAAUGACAUUUUCCAAACCAAUUUCCCCCCUGCCCUCCCUUCCACCAUGCCGCACCAUGAGGCACCACCAAAAGUGGA .(((((...((((((((....))))))))(((((((((....(((...................))).....))))...........((((......))))))))).........))))) ( -25.71) >consensus UUCCAUAUCAGUUGACAAAUUUGUCAACUUGCCUGGCAACUUGAAUGACAUUUUCCAAACCAAUUUCCCCCG______ACCAACCACC_________________ACCACCAAAAGUGGA .(((((...((((((((....))))))))....(((......(((.......)))....))).....................................................))))) (-12.46 = -12.68 + 0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:10 2006