
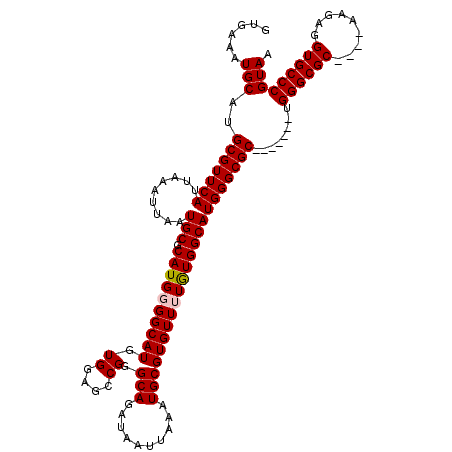
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,959,785 – 11,959,894 |
| Length | 109 |
| Max. P | 0.999943 |

| Location | 11,959,785 – 11,959,894 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.82 |
| Mean single sequence MFE | -38.97 |
| Consensus MFE | -30.56 |
| Energy contribution | -30.80 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
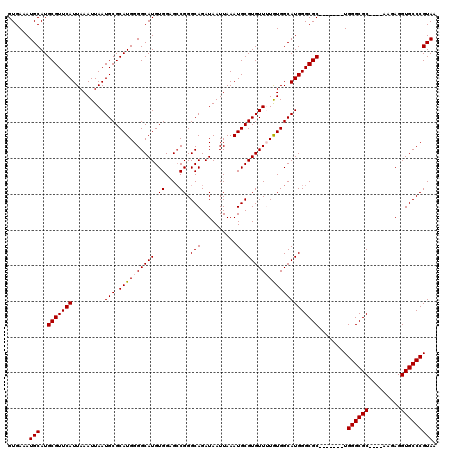
>2L_DroMel_CAF1 11959785 109 + 22407834 GUGAAAUGCAUGCGUUCAUUAAAUUAAUGCGCAUGGGGCAUGUGGAGCCGGGCAGAUAAUUAAAUGCGUGUUUUGUGGCAUGGGCGC-------UGGGCGC----AAGAGGUGCCCGUAA ((.(...(((((((((.....(((((.(((.(....(((.......)))).)))..))))).)))))))))....).))((((((((-------(...(..----..).))))))))).. ( -36.70) >DroSec_CAF1 179136 109 + 1 GUGAAAUGCAUGCGUUCAUUAAAUUAAUGCGCAUGGGGCAUGUGGUGCCGGGCAGAUAAUUAAAUGCGUGUUUUAUGGCAUGGGCGC-------UGGGCGC----AUGAGGUGCCCGUAA ........(((((((.............)))))))((((((.(.(((((.(((..((......)).((((((....))))))...))-------).)))))----...).)))))).... ( -38.32) >DroSim_CAF1 145585 109 + 1 GUGAAAUGCAUGCGUUCAUUAAAUUAAUGCGCAUGGGGCAUGUGGUGCCGGGCAGAUAAUUAAAUGCGUGUUUUGUGGCAUGGGCGC-------UGGGCGC----AUGAGGUGCCCGUAA ........(((((((.............)))))))((((((.(.(((((.(((..((......)).((((((....))))))...))-------).)))))----...).)))))).... ( -38.32) >DroEre_CAF1 154104 119 + 1 GUGAAAUGCAUGCGUUCAUUAAAUUAAUGCGCAUGGGGCAUGUGGAGACGGGCAGAUAAUUAAAUGCGUGUU-UGUGGCAUGGGCGCUACGCGCUGGGCGCUCGUAAGAGGUGCCCGUAA ......(.(((((((.............))))))).).(((((.((((((.(((..........))).))))-).).)))))(((((...)))))((((((((....)).)))))).... ( -44.12) >DroYak_CAF1 149916 108 + 1 GUGAAAUGCAUGCGUUCAUUAAAUUAAUGCGCAUGGGGCAUGUGGAGCCGGGCAGAUAAUUAAAUGCGUGUU-UGUGGCAUGGGCGC-------UGGGCGC----AAGAGGUGCCCGUAA ((.(...(((((((((.....(((((.(((.(....(((.......)))).)))..))))).))))))))).-..).))((((((((-------(...(..----..).))))))))).. ( -37.40) >consensus GUGAAAUGCAUGCGUUCAUUAAAUUAAUGCGCAUGGGGCAUGUGGAGCCGGGCAGAUAAUUAAAUGCGUGUUUUGUGGCAUGGGCGC_______UGGGCGC____AAGAGGUGCCCGUAA ......(((..(((((((.........(((.((((((((((.((....)).(((..........)))))))))))))))))))))))........((((((.........))))))))). (-30.56 = -30.80 + 0.24)

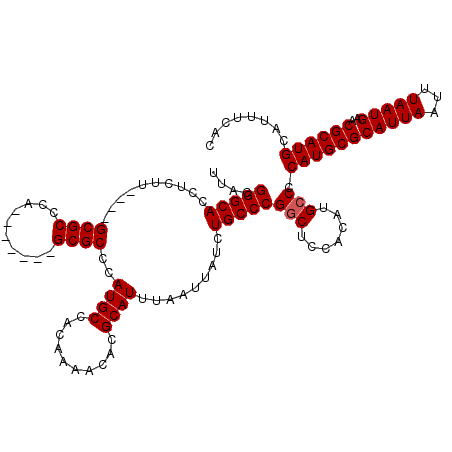
| Location | 11,959,785 – 11,959,894 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.82 |
| Mean single sequence MFE | -27.45 |
| Consensus MFE | -25.44 |
| Energy contribution | -25.64 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.73 |
| SVM RNA-class probability | 0.999943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
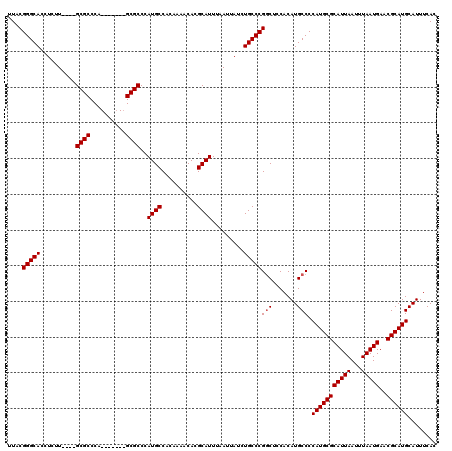
>2L_DroMel_CAF1 11959785 109 - 22407834 UUACGGGCACCUCUU----GCGCCCA-------GCGCCCAUGCCACAAAACACGCAUUUAAUUAUCUGCCCGGCUCCACAUGCCCCAUGCGCAUUAAUUUAAUGAACGCAUGCAUUUCAC ....(((((......----((((...-------))))..((((..........)))).........)))))(((.......))).(((((((((((...)))))..))))))........ ( -26.90) >DroSec_CAF1 179136 109 - 1 UUACGGGCACCUCAU----GCGCCCA-------GCGCCCAUGCCAUAAAACACGCAUUUAAUUAUCUGCCCGGCACCACAUGCCCCAUGCGCAUUAAUUUAAUGAACGCAUGCAUUUCAC ....(((((......----((((...-------))))..((((..........)))).........)))))((((.....)))).(((((((((((...)))))..))))))........ ( -27.40) >DroSim_CAF1 145585 109 - 1 UUACGGGCACCUCAU----GCGCCCA-------GCGCCCAUGCCACAAAACACGCAUUUAAUUAUCUGCCCGGCACCACAUGCCCCAUGCGCAUUAAUUUAAUGAACGCAUGCAUUUCAC ....(((((......----((((...-------))))..((((..........)))).........)))))((((.....)))).(((((((((((...)))))..))))))........ ( -27.40) >DroEre_CAF1 154104 119 - 1 UUACGGGCACCUCUUACGAGCGCCCAGCGCGUAGCGCCCAUGCCACA-AACACGCAUUUAAUUAUCUGCCCGUCUCCACAUGCCCCAUGCGCAUUAAUUUAAUGAACGCAUGCAUUUCAC ..(((((((.....((((.((.....)).))))(((...........-....)))...........)))))))............(((((((((((...)))))..))))))........ ( -28.56) >DroYak_CAF1 149916 108 - 1 UUACGGGCACCUCUU----GCGCCCA-------GCGCCCAUGCCACA-AACACGCAUUUAAUUAUCUGCCCGGCUCCACAUGCCCCAUGCGCAUUAAUUUAAUGAACGCAUGCAUUUCAC ....(((((......----((((...-------))))..((((....-.....)))).........)))))(((.......))).(((((((((((...)))))..))))))........ ( -27.00) >consensus UUACGGGCACCUCUU____GCGCCCA_______GCGCCCAUGCCACAAAACACGCAUUUAAUUAUCUGCCCGGCUCCACAUGCCCCAUGCGCAUUAAUUUAAUGAACGCAUGCAUUUCAC ....(((((..........((((..........))))..((((..........)))).........)))))(((.......))).(((((((((((...)))))..))))))........ (-25.44 = -25.64 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:08 2006