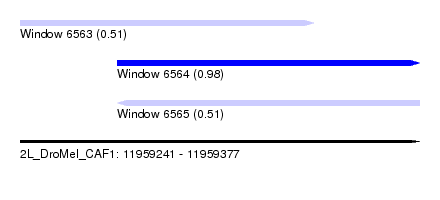
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,959,241 – 11,959,377 |
| Length | 136 |
| Max. P | 0.979545 |

| Location | 11,959,241 – 11,959,341 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.40 |
| Mean single sequence MFE | -22.27 |
| Consensus MFE | -7.40 |
| Energy contribution | -7.64 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.33 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11959241 100 + 22407834 GGCAGUAAUG-------AAAUGUCUGGCAUUUCGUCCGUUUCCAUCAGCUAACC---AACCAA----------CCAACUGCCAACUCCAAGCUGACCACUCUUAACGGCCAGCUAGCAAU ((((((.(((-------(((((.....))))))))..(((......))).....---......----------...)))))).......(((((.((.........)).)))))...... ( -24.90) >DroSec_CAF1 178616 92 + 1 GUCAGUAAUG-------AAAUGUCUGGCAUUUCGUCCGUUUCCAUCAGCUAAC---------------------CAACUGCCCACUUCAAGCUGACGACUCUCAACGGCCAGCUACCAAU ((((((..((-------((.((...((((.((.((..(((......)))..))---------------------.)).)))))).)))).))))))..........((.......))... ( -17.50) >DroSim_CAF1 145059 92 + 1 GUCAGUAAUG-------AAAUGUCUGGCAUUUCGUCCGUUUCCAUCAGCUAAC---------------------CAACUGCCCACUUCAAGCUGACGACUCUCAACGGCCAGCUACCAAU ((((((..((-------((.((...((((.((.((..(((......)))..))---------------------.)).)))))).)))).))))))..........((.......))... ( -17.50) >DroEre_CAF1 153578 84 + 1 GGCAGUAAUG-------AAAUGGCUGGCAUUUAGUCCGUUUCCAUCAGCUA-----CAGCUAC----------CCAACUGCCAACUCCAAG--------------CCGCCAGCUCCCAAU ((((((...(-------(((((((((.....))).)))))))....(((..-----..)))..----------...)))))).......((--------------(.....)))...... ( -21.80) >DroYak_CAF1 149339 111 + 1 GGCAGUAAUGUCGGGCAAAAUGUCCGGCAUUUCGUCCGUUUCCAUCAGCUAUCCAACAGCCAAGUGCCAACUGCCAACUGCUAACUUCAAA---------CUGAACUGCCAGCUCCCAAU ((((((((((((((((.....)))))))))).............((((.........(((..(((((.....))..)))))).........---------)))))))))).......... ( -29.67) >consensus GGCAGUAAUG_______AAAUGUCUGGCAUUUCGUCCGUUUCCAUCAGCUAAC____A_C_A___________CCAACUGCCAACUUCAAGCUGAC_ACUCUCAACGGCCAGCUACCAAU ((((((...........(((((.....))))).....(((......)))...........................))))))...................................... ( -7.40 = -7.64 + 0.24)

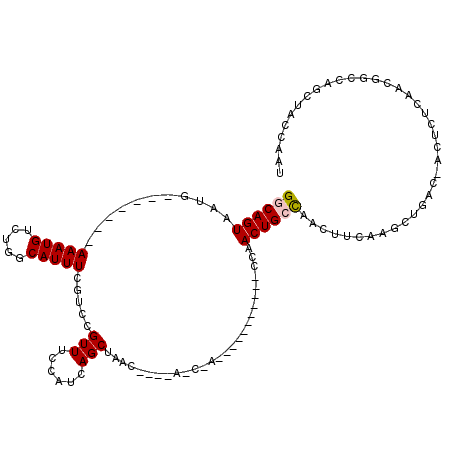
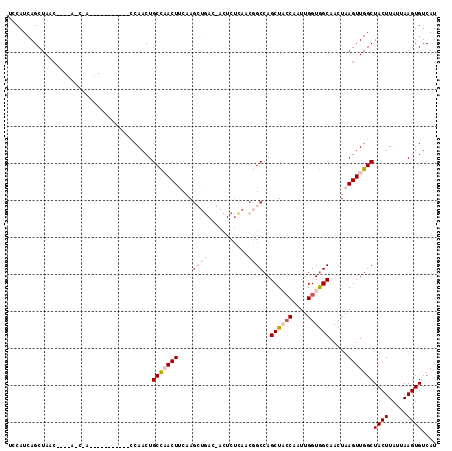
| Location | 11,959,274 – 11,959,377 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 77.39 |
| Mean single sequence MFE | -23.54 |
| Consensus MFE | -13.58 |
| Energy contribution | -15.66 |
| Covariance contribution | 2.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11959274 103 + 22407834 UCCAUCAGCUAACC---AACCAA----------CCAACUGCCAACUCCAAGCUGACCACUCUUAACGGCCAGCUAGCAAUUGGUGGCAACUAAGUUGGCUACUUAUUAAGUGUCAU ......(((((((.---.....(----------((((.(((........(((((.((.........)).))))).))).)))))(....)...)))))))................ ( -23.50) >DroSec_CAF1 178649 95 + 1 UCCAUCAGCUAAC---------------------CAACUGCCCACUUCAAGCUGACGACUCUCAACGGCCAGCUACCAAUUGGUGGCAACUAAGUUGGCUACUUAUUAAGUGUCAU ....((((((...---------------------...............)))))).(((.......(((((((((((....)))(....)..))))))))(((.....)))))).. ( -23.47) >DroSim_CAF1 145092 95 + 1 UCCAUCAGCUAAC---------------------CAACUGCCCACUUCAAGCUGACGACUCUCAACGGCCAGCUACCAAUUGGUGGCAACUAAGUUGGCUACUUAUUAAGUGUCAU ....((((((...---------------------...............)))))).(((.......(((((((((((....)))(....)..))))))))(((.....)))))).. ( -23.47) >DroEre_CAF1 153611 87 + 1 UCCAUCAGCUA-----CAGCUAC----------CCAACUGCCAACUCCAAG--------------CCGCCAGCUCCCAAUUGGUGGCAACUAAGUUGGCUACUUAUUAAGUGUCAU ......(((..-----..)))..----------......(((((((....(--------------(((((((.......)))))))).....)))))))((((.....)))).... ( -24.70) >DroYak_CAF1 149379 107 + 1 UCCAUCAGCUAUCCAACAGCCAAGUGCCAACUGCCAACUGCUAACUUCAAA---------CUGAACUGCCAGCUCCCAAUUGGUAGCAACUAAGUUGGCUACUUAUUAAGUGUCAU .......(((.......))).(((((((((((.....(((((((.......---------(((......))).......)))))))......))))))).))))............ ( -22.54) >consensus UCCAUCAGCUAAC____A_C_A___________CCAACUGCCAACUUCAAGCUGAC_ACUCUCAACGGCCAGCUACCAAUUGGUGGCAACUAAGUUGGCUACUUAUUAAGUGUCAU .......................................(((((((....((((...........))))..((((((....)))))).....)))))))((((.....)))).... (-13.58 = -15.66 + 2.08)

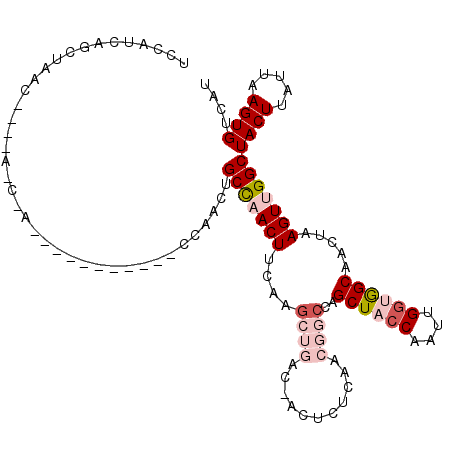
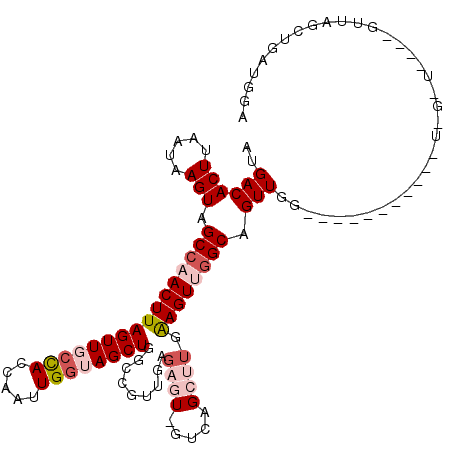
| Location | 11,959,274 – 11,959,377 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 77.39 |
| Mean single sequence MFE | -30.74 |
| Consensus MFE | -14.04 |
| Energy contribution | -15.94 |
| Covariance contribution | 1.90 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.46 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11959274 103 - 22407834 AUGACACUUAAUAAGUAGCCAACUUAGUUGCCACCAAUUGCUAGCUGGCCGUUAAGAGUGGUCAGCUUGGAGUUGGCAGUUGG----------UUGGUU---GGUUAGCUGAUGGA ....((.(((.(((.(((((((((...((((((...(((.(.(((((((((.......))))))))).).)))))))))..))----------))))))---).)))..))))).. ( -37.80) >DroSec_CAF1 178649 95 - 1 AUGACACUUAAUAAGUAGCCAACUUAGUUGCCACCAAUUGGUAGCUGGCCGUUGAGAGUCGUCAGCUUGAAGUGGGCAGUUG---------------------GUUAGCUGAUGGA ......((.....(((((((((((.((((((((.....))))))))(.(((((..((((.....))))..))))).))))))---------------------))).)))...)). ( -28.30) >DroSim_CAF1 145092 95 - 1 AUGACACUUAAUAAGUAGCCAACUUAGUUGCCACCAAUUGGUAGCUGGCCGUUGAGAGUCGUCAGCUUGAAGUGGGCAGUUG---------------------GUUAGCUGAUGGA ......((.....(((((((((((.((((((((.....))))))))(.(((((..((((.....))))..))))).))))))---------------------))).)))...)). ( -28.30) >DroEre_CAF1 153611 87 - 1 AUGACACUUAAUAAGUAGCCAACUUAGUUGCCACCAAUUGGGAGCUGGCGG--------------CUUGGAGUUGGCAGUUGG----------GUAGCUG-----UAGCUGAUGGA .....(((((((.....((((((((((((((((((.....))...))))))--------------))..)))))))).)))))----------))(((..-----..)))...... ( -28.10) >DroYak_CAF1 149379 107 - 1 AUGACACUUAAUAAGUAGCCAACUUAGUUGCUACCAAUUGGGAGCUGGCAGUUCAG---------UUUGAAGUUAGCAGUUGGCAGUUGGCACUUGGCUGUUGGAUAGCUGAUGGA ...........(((((.(((((((((..(((((((....))(((((((....))))---------))).....)))))..))..)))))))))))).((((..(....)..)))). ( -31.20) >consensus AUGACACUUAAUAAGUAGCCAACUUAGUUGCCACCAAUUGGUAGCUGGCCGUUGAGAGU_GUCAGCUUGAAGUUGGCAGUUGG___________U_G_U____GUUAGCUGAUGGA ..((((((.....))).((((((((((((((((.....)))))))).........((((.....)))).)))))))).)))................................... (-14.04 = -15.94 + 1.90)

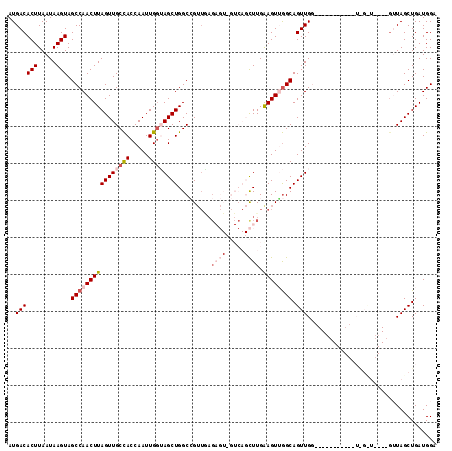
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:06 2006