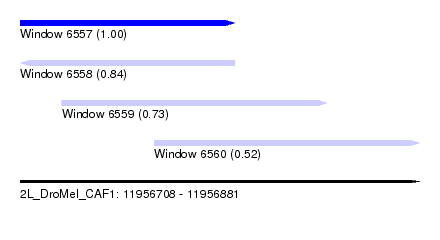
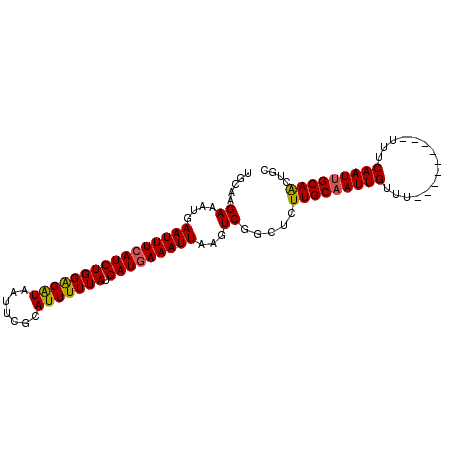
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,956,708 – 11,956,881 |
| Length | 173 |
| Max. P | 0.998004 |

| Location | 11,956,708 – 11,956,801 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 89.66 |
| Mean single sequence MFE | -21.30 |
| Consensus MFE | -18.92 |
| Energy contribution | -18.84 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.998004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11956708 93 + 22407834 UGCAAGAAAUGAAUUUCAUCUGGAGAUAAUUCGCAUUUUUAUGAUGAAAUUAAGUCGACUCUUGCAAUUGUUUUU-----UUUUUCAAUUGCAGCUGG .((..((....((((((((((((((((.......))))))).)))))))))...))......((((((((.....-----.....))))))))))... ( -22.70) >DroSec_CAF1 176075 89 + 1 UGCAAGAAAUGAAUUUCAUCUGGAGAUAAUUCACAUUUUUAUGAUGAAAUUAAGUCGGGUCUUGCAAUUGUUU---------UUUCAAUUGCAACUGG .((..((....((((((((((((((((.......))))))).)))))))))...))..)).(((((((((...---------...))))))))).... ( -23.50) >DroSim_CAF1 142474 89 + 1 UGCAAGAAAUGAAUUUCAUCUGGGGGUAAUUCACAUUUUUAUGAUGAAAUUAAGUCGGCUCUUGCAAUUGUUU---------UUUCAAUUGCAACUGA .((..((....((((((((((((((((.......))))))).)))))))))...)).))..(((((((((...---------...))))))))).... ( -22.00) >DroEre_CAF1 151005 89 + 1 UGCAAGAAAUGAAUUUCAUCUGGAGAUAAUUCGCAUUUUUAUGAUGAAAUUAAGUCCGCUCUUGCCAUUGUUA---------UUUCAAUUGCAACUGC .....((....((((((((((((((((.......))))))).)))))))))...)).((..((((.((((...---------...)))).))))..)) ( -20.10) >DroYak_CAF1 146823 98 + 1 UGCAAGAAAUGAAUUUCAUCUGGAGAUAAUUCGCAUUUUUAUGAUCAAAUUAAGUCAGCUCUUGCAAUUGUUUUUUUUUUUUUUUCAAUUGCAACUGC (((((....((((........((((((((((.(((......((((........)))).....)))))))))))))........)))).)))))..... ( -18.19) >consensus UGCAAGAAAUGAAUUUCAUCUGGAGAUAAUUCGCAUUUUUAUGAUGAAAUUAAGUCGGCUCUUGCAAUUGUUU_________UUUCAAUUGCAACUGC .....((....((((((((((((((((.......))))))).)))))))))...)).....(((((((((...............))))))))).... (-18.92 = -18.84 + -0.08)

| Location | 11,956,708 – 11,956,801 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 89.66 |
| Mean single sequence MFE | -16.86 |
| Consensus MFE | -12.53 |
| Energy contribution | -12.93 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841302 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11956708 93 - 22407834 CCAGCUGCAAUUGAAAAA-----AAAAACAAUUGCAAGAGUCGACUUAAUUUCAUCAUAAAAAUGCGAAUUAUCUCCAGAUGAAAUUCAUUUCUUGCA ...((.(((((((.....-----.....)))))))(((((..((....((((((((......................))))))))))..))))))). ( -16.75) >DroSec_CAF1 176075 89 - 1 CCAGUUGCAAUUGAAA---------AAACAAUUGCAAGACCCGACUUAAUUUCAUCAUAAAAAUGUGAAUUAUCUCCAGAUGAAAUUCAUUUCUUGCA ....(((((((((...---------...))))))))).....((..((((((((((......................))))))))).)..))..... ( -15.65) >DroSim_CAF1 142474 89 - 1 UCAGUUGCAAUUGAAA---------AAACAAUUGCAAGAGCCGACUUAAUUUCAUCAUAAAAAUGUGAAUUACCCCCAGAUGAAAUUCAUUUCUUGCA ....(((((((((...---------...)))))))))..((.((..((((((((((......................))))))))).)..))..)). ( -17.25) >DroEre_CAF1 151005 89 - 1 GCAGUUGCAAUUGAAA---------UAACAAUGGCAAGAGCGGACUUAAUUUCAUCAUAAAAAUGCGAAUUAUCUCCAGAUGAAAUUCAUUUCUUGCA ((....)).((((...---------...)))).((((((..(((..(((((((((.......))).))))))..))).((((.....)))))))))). ( -18.00) >DroYak_CAF1 146823 98 - 1 GCAGUUGCAAUUGAAAAAAAAAAAAAAACAAUUGCAAGAGCUGACUUAAUUUGAUCAUAAAAAUGCGAAUUAUCUCCAGAUGAAAUUCAUUUCUUGCA (((((((((((((...............))))))))).....((..(((((((..(((....))))))))))..)).(((((.....))))).)))). ( -16.66) >consensus CCAGUUGCAAUUGAAA_________AAACAAUUGCAAGAGCCGACUUAAUUUCAUCAUAAAAAUGCGAAUUAUCUCCAGAUGAAAUUCAUUUCUUGCA .................................(((((((..((....((((((((......................))))))))))..))))))). (-12.53 = -12.93 + 0.40)

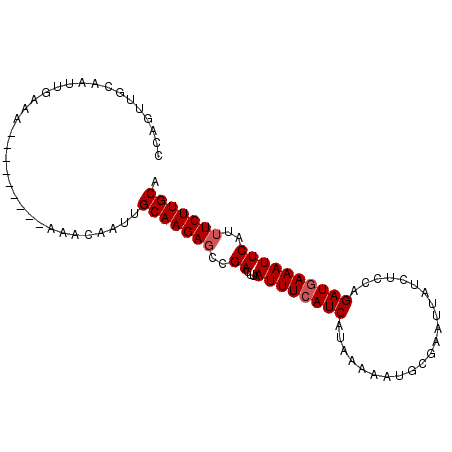
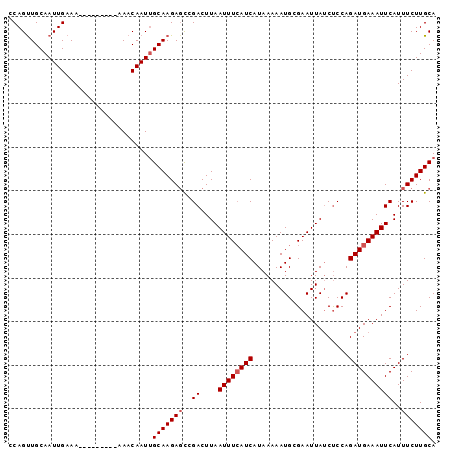
| Location | 11,956,726 – 11,956,841 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.19 |
| Mean single sequence MFE | -24.99 |
| Consensus MFE | -16.91 |
| Energy contribution | -17.91 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725687 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11956726 115 + 22407834 UCUGGAGAUAAUUCGCAUUUUUAUGAUGAAAUUAAGUCGACUCUUGCAAUUGUUUUU-----UUUUUCAAUUGCAGCUGGCAAGCUAGUGUAUAUAAACCUACUUGGGUGUGCCACUAUU ...(..(((......((((.....)))).......)))..)..(((((((((.....-----.....))))))))).(((((.(((...(((........)))...))).)))))..... ( -23.02) >DroSec_CAF1 176093 106 + 1 UCUGGAGAUAAUUCACAUUUUUAUGAUGAAAUUAAGUCGGGUCUUGCAAUUGUUU---------UUUCAAUUGCAACUGGCAAGCUA---UAU--AUAACAACUUGGGUGUGCCACUAUU .((.((..((((((((((....))).)).)))))..)).))..(((((((((...---------...))))))))).(((((.(((.---...--...........))).)))))..... ( -23.06) >DroSim_CAF1 142492 108 + 1 UCUGGGGGUAAUUCACAUUUUUAUGAUGAAAUUAAGUCGGCUCUUGCAAUUGUUU---------UUUCAAUUGCAACUGACAAGCUA---UAUAUAUACCGACUUGGGUGUGCCACUAUU ..(((.((((.(((((((....))).)))).(((((((((...(((((((((...---------...)))))))))((....))...---........)))))))))...)))).))).. ( -28.60) >DroYak_CAF1 146841 111 + 1 UCUGGAGAUAAUUCGCAUUUUUAUGAUCAAAUUAAGUCAGCUCUUGCAAUUGUUUUUUUUUUUUUUUCAAUUGCAACUGCCAAUCUU---UAU----ACCCACAUGGGUGUGCCAAUC-- ..(((((((..............((((........))))((..(((((((((...............)))))))))..))..)))).---(((----((((....))))))))))...-- ( -25.26) >consensus UCUGGAGAUAAUUCACAUUUUUAUGAUGAAAUUAAGUCGGCUCUUGCAAUUGUUU_________UUUCAAUUGCAACUGGCAAGCUA___UAU__AUACCAACUUGGGUGUGCCACUAUU ..(((......(((((((....))).)))).....(((((...(((((((((...............))))))))))))))..............(((((......))))).)))..... (-16.91 = -17.91 + 1.00)



| Location | 11,956,766 – 11,956,881 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.42 |
| Mean single sequence MFE | -24.06 |
| Consensus MFE | -12.50 |
| Energy contribution | -14.06 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.52 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11956766 115 + 22407834 CUCUUGCAAUUGUUUUU-----UUUUUCAAUUGCAGCUGGCAAGCUAGUGUAUAUAAACCUACUUGGGUGUGCCACUAUUGAGUGAACCAUUGAUUUAAGAAAUAAAGCCUUUAUUUCUG ((((((((((((.....-----.....))))))))).(((((.(((...(((........)))...))).))))).....)))...............((((((((.....)))))))). ( -26.00) >DroSec_CAF1 176133 106 + 1 GUCUUGCAAUUGUUU---------UUUCAAUUGCAACUGGCAAGCUA---UAU--AUAACAACUUGGGUGUGCCACUAUUGAGUGCACCAUUGAUUUAAGAAAUAAAGCCUUUAUUUCUG ((((((((((((...---------...)))))))))..)))......---...--...........((((..(((....)).)..)))).........((((((((.....)))))))). ( -27.20) >DroSim_CAF1 142532 108 + 1 CUCUUGCAAUUGUUU---------UUUCAAUUGCAACUGACAAGCUA---UAUAUAUACCGACUUGGGUGUGCCACUAUUGAGUGAACCAUUGAUUUAAGAAAUAAAGCCUUUAUUUCUG (..(((((((((...---------...)))))))))..).(((....---....((((((......)))))).((((....)))).....))).....((((((((.....)))))))). ( -23.40) >DroYak_CAF1 146881 109 + 1 CUCUUGCAAUUGUUUUUUUUUUUUUUUCAAUUGCAACUGCCAAUCUU---UAU----ACCCACAUGGGUGUGCCAAUC----GUUAUCCACUGAUUUAAGACAUAAAUCUGCCUUUCCUA ...(((((((((...............)))))))))..((.......---(((----((((....)))))))......----..........((((((.....)))))).))........ ( -19.66) >consensus CUCUUGCAAUUGUUU_________UUUCAAUUGCAACUGGCAAGCUA___UAU__AUACCAACUUGGGUGUGCCACUAUUGAGUGAACCAUUGAUUUAAGAAAUAAAGCCUUUAUUUCUG ...(((((((((...............))))))))).(((((..((....................))..))))).......................((((((((.....)))))))). (-12.50 = -14.06 + 1.56)

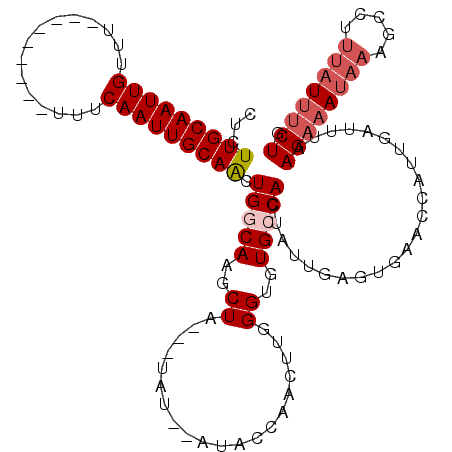

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:01 2006