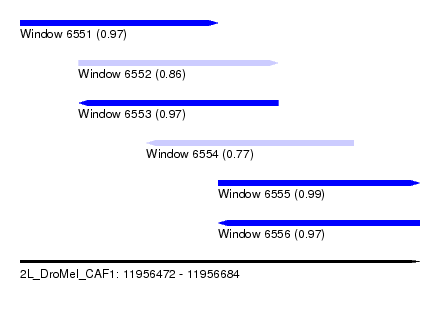
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,956,472 – 11,956,684 |
| Length | 212 |
| Max. P | 0.985144 |

| Location | 11,956,472 – 11,956,577 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 95.05 |
| Mean single sequence MFE | -33.48 |
| Consensus MFE | -30.14 |
| Energy contribution | -31.70 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11956472 105 + 22407834 CUGAUCUUUCCCCUC--CCGUUUUCGUUACGUUUCGUUUCGUGCUCUGAAAAUUGUUUUCGACGAAAAUUGUUGCACGAACCGAAAUGCAACAGAGGUG--GAGAUAUC ..(((.(((((((((--..(....)(((.(((((((.(((((((...((((.....))))(((((...)))))))))))).))))))).))).)))).)--)))).))) ( -34.40) >DroSec_CAF1 175837 107 + 1 CUGAUCUUUCCCCUC--CCGUUUUCGUUACGAUUCGUUUCGUGCUCUGAAAAUUGUUUUCGACGAAAAUUGUUGCACGAACCGAAAUGCAGCAGAGGUGGGGAGAUAUC ...((((..((((((--..(((((((((((((......)))).....((((.....)))))))))))))(((((((..........))))))))))).))..))))... ( -35.50) >DroSim_CAF1 142235 107 + 1 CUGAUCUUUCCCCUC--CCGUUUUCGUUACGUUUCGUUUCGUGCUCUGAAAAUUGUUUUCGACGAAAAUUGUUGCACGAACCGAAAUGCAGCAGAGGUGGGGAGAUAUC ...((((..((((((--..(....)(((.(((((((.(((((((...((((.....))))(((((...)))))))))))).))))))).))).)))).))..))))... ( -38.00) >DroEre_CAF1 150771 107 + 1 CUGAUCUUUUCCCCCCGCCGUUUUCGUUACGUUUCGUUUCAUGCUCUGAAAAUUGUUUUCGACGAAAAUUGUUGCACGAACCGAAAUGCAGCAGUGGUG--GAGAUAUC ..(((..(((((..((((.(((((((((........(((((.....))))).........)))))))))(((((((..........))))))))))).)--)))).))) ( -26.03) >consensus CUGAUCUUUCCCCUC__CCGUUUUCGUUACGUUUCGUUUCGUGCUCUGAAAAUUGUUUUCGACGAAAAUUGUUGCACGAACCGAAAUGCAGCAGAGGUG__GAGAUAUC ...((((((((((((....(....)(((.(((((((.(((((((...((((.....))))(((((...)))))))))))).))))))).))).)))).))))))))... (-30.14 = -31.70 + 1.56)


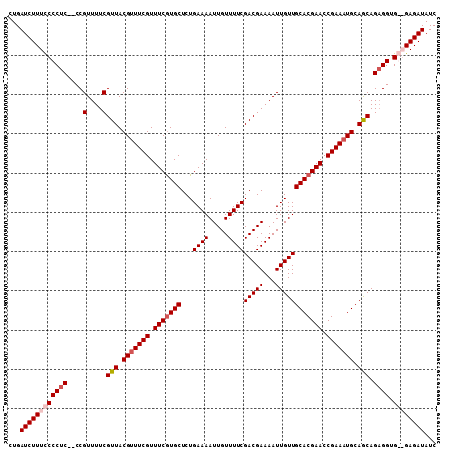
| Location | 11,956,503 – 11,956,609 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 91.49 |
| Mean single sequence MFE | -29.29 |
| Consensus MFE | -23.19 |
| Energy contribution | -23.59 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11956503 106 + 22407834 UCGUUUCGUGCUCUGAAAAUUGUUUUCGACGAAAAUUGUUGCACGAACCGAAAUGCAACAGAGGUG--GAGAUAUCAAUAUGGAUAUCUGU--------AUAUCUGUAUGGUCUUU (((.(((((((...((((.....))))(((((...)))))))))))).)))....(((((((.(((--.(((((((......))))))).)--------)).))))).))...... ( -32.10) >DroSec_CAF1 175868 108 + 1 UCGUUUCGUGCUCUGAAAAUUGUUUUCGACGAAAAUUGUUGCACGAACCGAAAUGCAGCAGAGGUGGGGAGAUAUCAAUAUGGAUAUCUGU--------AUAUCUGUAUGGUAUGU (((.(((((((...((((.....))))(((((...)))))))))))).))).((((.(((((.....(.(((((((......))))))).)--------...)))))...)))).. ( -31.30) >DroSim_CAF1 142266 108 + 1 UCGUUUCGUGCUCUGAAAAUUGUUUUCGACGAAAAUUGUUGCACGAACCGAAAUGCAGCAGAGGUGGGGAGAUAUCAAUAUGGAUAUCUGU--------GUAUCUGUAUGGUCUGU (((.(((((((...((((.....))))(((((...)))))))))))).)))...((((((.(((((.(.(((((((......))))))).)--------.))))).)...).)))) ( -31.00) >DroEre_CAF1 150804 103 + 1 UCGUUUCAUGCUCUGAAAAUUGUUUUCGACGAAAAUUGUUGCACGAACCGAAAUGCAGCAGUGGUG--GAGAUAUCAAUAUGGAUAUCUGU--------AAAUCUGUAUGGGC--- (((.(((.(((...((((.....))))(((((...)))))))).))).)))......((((...((--.(((((((......))))))).)--------)...))))......--- ( -23.50) >DroYak_CAF1 146613 112 + 1 UCGUUUCGUGCUCUGAAAAUUGUUUUCGACGAAAAUUGUUGCACGAACCGAAAUGCAGCUGAGGUG--GAGAUAUCAAUAUGGAUAUCUGUAUAAAUGUAUAUCUGUAUGGUCU-- (((.(((((((...((((.....))))(((((...)))))))))))).))).((((((.....(((--.(((((((......))))))).)))..........)))))).....-- ( -28.56) >consensus UCGUUUCGUGCUCUGAAAAUUGUUUUCGACGAAAAUUGUUGCACGAACCGAAAUGCAGCAGAGGUG__GAGAUAUCAAUAUGGAUAUCUGU________AUAUCUGUAUGGUCU_U (((.(((((((...((((.....))))(((((...)))))))))))).))).((((((...........(((((((......)))))))..............))))))....... (-23.19 = -23.59 + 0.40)

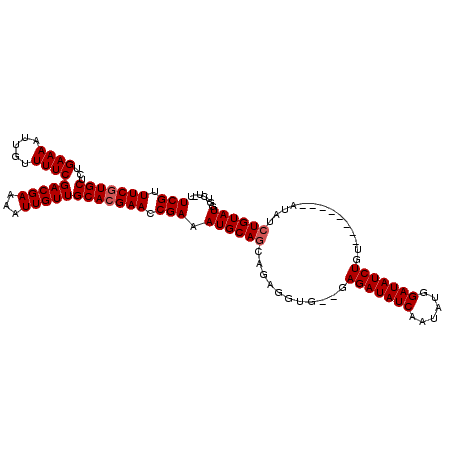
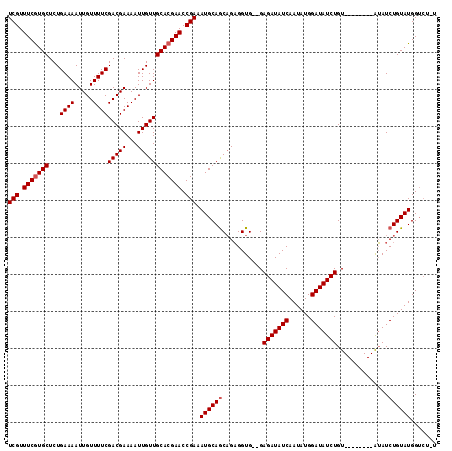
| Location | 11,956,503 – 11,956,609 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 91.49 |
| Mean single sequence MFE | -20.58 |
| Consensus MFE | -18.53 |
| Energy contribution | -18.21 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11956503 106 - 22407834 AAAGACCAUACAGAUAU--------ACAGAUAUCCAUAUUGAUAUCUC--CACCUCUGUUGCAUUUCGGUUCGUGCAACAAUUUUCGUCGAAAACAAUUUUCAGAGCACGAAACGA ......((.(((((...--------..(((((((......))))))).--....)))))))....(((.(((((((.((.......)).((((.....))))...))))))).))) ( -23.30) >DroSec_CAF1 175868 108 - 1 ACAUACCAUACAGAUAU--------ACAGAUAUCCAUAUUGAUAUCUCCCCACCUCUGCUGCAUUUCGGUUCGUGCAACAAUUUUCGUCGAAAACAAUUUUCAGAGCACGAAACGA ..........((((...--------..(((((((......))))))).......)))).......(((.(((((((.((.......)).((((.....))))...))))))).))) ( -20.90) >DroSim_CAF1 142266 108 - 1 ACAGACCAUACAGAUAC--------ACAGAUAUCCAUAUUGAUAUCUCCCCACCUCUGCUGCAUUUCGGUUCGUGCAACAAUUUUCGUCGAAAACAAUUUUCAGAGCACGAAACGA ..........((((...--------..(((((((......))))))).......)))).......(((.(((((((.((.......)).((((.....))))...))))))).))) ( -20.90) >DroEre_CAF1 150804 103 - 1 ---GCCCAUACAGAUUU--------ACAGAUAUCCAUAUUGAUAUCUC--CACCACUGCUGCAUUUCGGUUCGUGCAACAAUUUUCGUCGAAAACAAUUUUCAGAGCAUGAAACGA ---((.....(((....--------..(((((((......))))))).--.....)))..))...(((.(((((((.((.......)).((((.....))))...))))))).))) ( -18.90) >DroYak_CAF1 146613 112 - 1 --AGACCAUACAGAUAUACAUUUAUACAGAUAUCCAUAUUGAUAUCUC--CACCUCAGCUGCAUUUCGGUUCGUGCAACAAUUUUCGUCGAAAACAAUUUUCAGAGCACGAAACGA --........((((((......)))..(((((((......))))))).--........)))....(((.(((((((.((.......)).((((.....))))...))))))).))) ( -18.90) >consensus A_AGACCAUACAGAUAU________ACAGAUAUCCAUAUUGAUAUCUC__CACCUCUGCUGCAUUUCGGUUCGUGCAACAAUUUUCGUCGAAAACAAUUUUCAGAGCACGAAACGA ..........(((..............(((((((......)))))))...........)))....(((.(((((((.((.......)).((((.....))))...))))))).))) (-18.53 = -18.21 + -0.32)



| Location | 11,956,539 – 11,956,649 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.85 |
| Mean single sequence MFE | -27.29 |
| Consensus MFE | -23.22 |
| Energy contribution | -23.22 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768401 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11956539 110 - 22407834 CUUACAUGCCCGCAAAUGUAUGUAGGAAUGCUGGUGGCAUAAAGACCAUACAGAUAU--------ACAGAUAUCCAUAUUGAUAUCUC--CACCUCUGUUGCAUUUCGGUUCGUGCAACA (((((((((........)))))))))......(((((....................--------..(((((((......))))))))--))))...(((((((........))))))). ( -29.14) >DroSec_CAF1 175904 112 - 1 CUUAUAUGCCAGCAAAUGUAUGUAGGAAUGCUGGUGGCAUACAUACCAUACAGAUAU--------ACAGAUAUCCAUAUUGAUAUCUCCCCACCUCUGCUGCAUUUCGGUUCGUGCAACA ................((((((..((((((((((((.......)))))..((((...--------..(((((((......))))))).......))))..)))))))....))))))... ( -26.70) >DroSim_CAF1 142302 112 - 1 CUUAUAUGCCAGCAAAUGUAUGUAGGAAUGCUGGUGGCAUACAGACCAUACAGAUAC--------ACAGAUAUCCAUAUUGAUAUCUCCCCACCUCUGCUGCAUUUCGGUUCGUGCAACA ................((((((..(((((((..((((........)))).((((...--------..(((((((......))))))).......))))..)))))))....))))))... ( -25.80) >DroEre_CAF1 150840 106 - 1 CGUACAUGGCAGCAAAUGUAUGUAGGAAUGCUGGUGGCA----GCCCAUACAGAUUU--------ACAGAUAUCCAUAUUGAUAUCUC--CACCACUGCUGCAUUUCGGUUCGUGCAACA .((((..(((.(.(((((((.((((((((.(((((((..----..)))).)))))))--------..(((((((......))))))).--.....)))))))))))).))).)))).... ( -27.50) >DroYak_CAF1 146649 115 - 1 CUUACAUGGCAGCAAAUGUAUGUAGGAAUGCUGGUGGCA---AGACCAUACAGAUAUACAUUUAUACAGAUAUCCAUAUUGAUAUCUC--CACCUCAGCUGCAUUUCGGUUCGUGCAACA ........(((((((((((((((.......(((((((..---...)))).)))))))))))))....(((((((......))))))).--.......))))).................. ( -27.31) >consensus CUUACAUGCCAGCAAAUGUAUGUAGGAAUGCUGGUGGCAUA_AGACCAUACAGAUAU________ACAGAUAUCCAUAUUGAUAUCUC__CACCUCUGCUGCAUUUCGGUUCGUGCAACA ................((((((..(((((((..((((........))))..................(((((((......))))))).............)))))))....))))))... (-23.22 = -23.22 + 0.00)

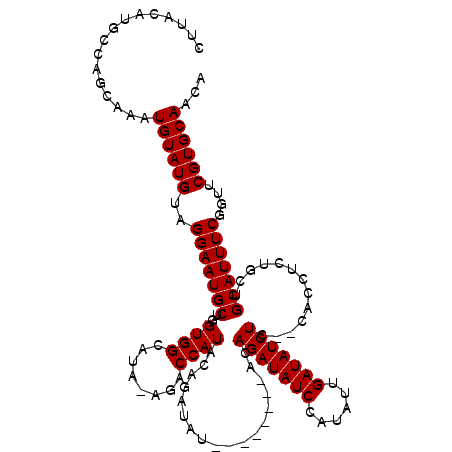
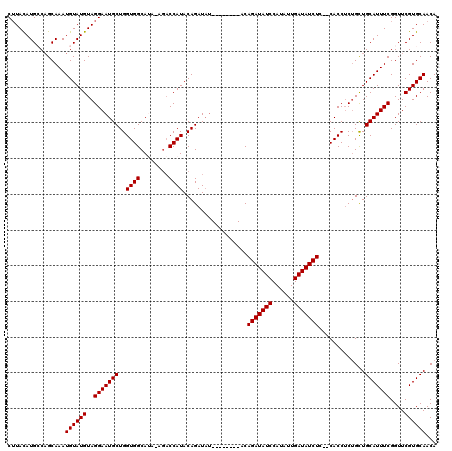
| Location | 11,956,577 – 11,956,684 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 90.90 |
| Mean single sequence MFE | -28.68 |
| Consensus MFE | -22.26 |
| Energy contribution | -23.06 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11956577 107 + 22407834 AAUAUGGAUAUCUGU--------AUAUCUGUAUGGUCUUUAUGCCACCAGCAUUCCUACAUACAUUUGCGGGCAUGUAAGUUGAUUCUUAUUAACCAUAUAUAGGUCAGCAGCCU .((((((((((....--------))))))))))(((...((((((....(((..............))).))))))...(((((((..(((.......)))..))))))).))). ( -26.04) >DroSec_CAF1 175944 107 + 1 AAUAUGGAUAUCUGU--------AUAUCUGUAUGGUAUGUAUGCCACCAGCAUUCCUACAUACAUUUGCUGGCAUAUAAGUUGGUUCUUAUUAACCAUAUAUAGGUCAGCAGCCU .((((((((((....--------)))))))))).((((((((((.....)))....)))))))..((((((((.((((...(((((......)))))..)))).))))))))... ( -31.80) >DroSim_CAF1 142342 107 + 1 AAUAUGGAUAUCUGU--------GUAUCUGUAUGGUCUGUAUGCCACCAGCAUUCCUACAUACAUUUGCUGGCAUAUAAGUUGGUUCUUAUUAACCAUAUAUAGGUCAGCAGCCU .((((((((((....--------))))))))))((.((((..(((.((((((..............))))))..((((...(((((......)))))..)))))))..)))))). ( -30.04) >DroEre_CAF1 150878 103 + 1 AAUAUGGAUAUCUGU--------AAAUCUGUAUGGGC----UGCCACCAGCAUUCCUACAUACAUUUGCUGCCAUGUACGUUGGUUCUUAUUAGCCAUAUAUAGGUCAGCAGCCU .((((((((......--------..))))))))((((----(((...(((((..............)))))((.((((...(((((......)))))..))))))...))))))) ( -27.84) >DroYak_CAF1 146687 112 + 1 AAUAUGGAUAUCUGUAUAAAUGUAUAUCUGUAUGGUCU---UGCCACCAGCAUUCCUACAUACAUUUGCUGCCAUGUAAGUUGGUUCUUAUUAACCAUAUAUAGGUCAGCAGCCU .((((((....))))))(((((((....((((.((...---(((.....)))..)))))))))))))((((((.((((...(((((......)))))..)))))).))))..... ( -27.70) >consensus AAUAUGGAUAUCUGU________AUAUCUGUAUGGUCU_UAUGCCACCAGCAUUCCUACAUACAUUUGCUGGCAUGUAAGUUGGUUCUUAUUAACCAUAUAUAGGUCAGCAGCCU .....((.....................((((((......((((.....)))).....)))))).((((((((.((((...(((((......)))))..)))).)))))))))). (-22.26 = -23.06 + 0.80)

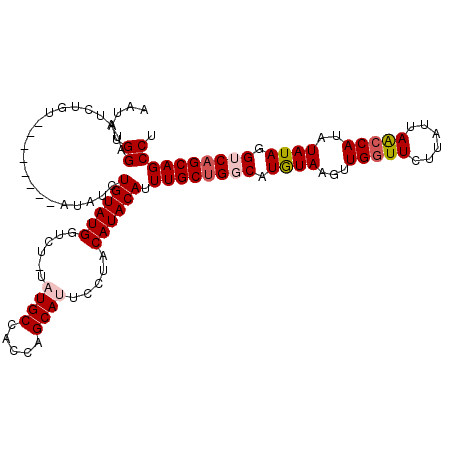
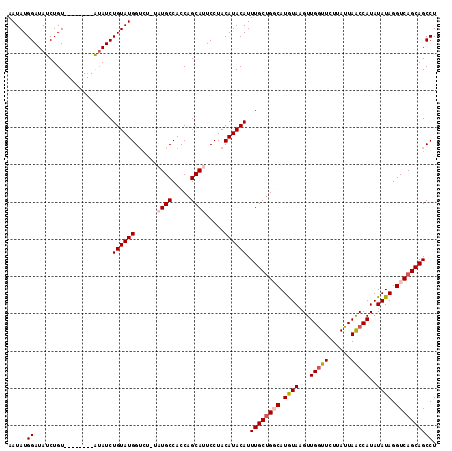
| Location | 11,956,577 – 11,956,684 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 90.90 |
| Mean single sequence MFE | -25.48 |
| Consensus MFE | -22.32 |
| Energy contribution | -22.12 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11956577 107 - 22407834 AGGCUGCUGACCUAUAUAUGGUUAAUAAGAAUCAACUUACAUGCCCGCAAAUGUAUGUAGGAAUGCUGGUGGCAUAAAGACCAUACAGAUAU--------ACAGAUAUCCAUAUU .((.(((((...(((.(((((((............(((((((((........))))))))).((((.....))))...)))))))...))).--------.))).)).))..... ( -24.60) >DroSec_CAF1 175944 107 - 1 AGGCUGCUGACCUAUAUAUGGUUAAUAAGAACCAACUUAUAUGCCAGCAAAUGUAUGUAGGAAUGCUGGUGGCAUACAUACCAUACAGAUAU--------ACAGAUAUCCAUAUU .((.(((((.(.((((..(((((......)))))...)))).).))))).((((((((.((....))....)))))))).)).....(((((--------....)))))...... ( -24.60) >DroSim_CAF1 142342 107 - 1 AGGCUGCUGACCUAUAUAUGGUUAAUAAGAACCAACUUAUAUGCCAGCAAAUGUAUGUAGGAAUGCUGGUGGCAUACAGACCAUACAGAUAC--------ACAGAUAUCCAUAUU .((.(((((...(((.(((((((..((((......))))(((((((.((...((((......)))))).)))))))..)))))))...))).--------.))).)).))..... ( -25.60) >DroEre_CAF1 150878 103 - 1 AGGCUGCUGACCUAUAUAUGGCUAAUAAGAACCAACGUACAUGGCAGCAAAUGUAUGUAGGAAUGCUGGUGGCA----GCCCAUACAGAUUU--------ACAGAUAUCCAUAUU .(((((((.(((..(((....((....))..((.((((((((........)))))))).)).)))..)))))))----)))...........--------............... ( -26.80) >DroYak_CAF1 146687 112 - 1 AGGCUGCUGACCUAUAUAUGGUUAAUAAGAACCAACUUACAUGGCAGCAAAUGUAUGUAGGAAUGCUGGUGGCA---AGACCAUACAGAUAUACAUUUAUACAGAUAUCCAUAUU ..((((((((((.......))))..((((......))))...))))))((((((((((.......(((((((..---...)))).)))))))))))))................. ( -25.81) >consensus AGGCUGCUGACCUAUAUAUGGUUAAUAAGAACCAACUUACAUGCCAGCAAAUGUAUGUAGGAAUGCUGGUGGCAUA_AGACCAUACAGAUAU________ACAGAUAUCCAUAUU .((.(((((.((((((..(((((......)))))...(((((........)))))))))))....(((((((........)))).))).............))).)).))..... (-22.32 = -22.12 + -0.20)

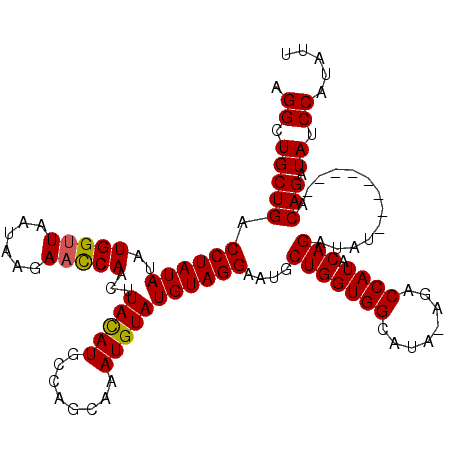
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:58 2006