
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,956,280 – 11,956,409 |
| Length | 129 |
| Max. P | 0.998817 |

| Location | 11,956,280 – 11,956,380 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 97.80 |
| Mean single sequence MFE | -11.25 |
| Consensus MFE | -10.84 |
| Energy contribution | -10.73 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
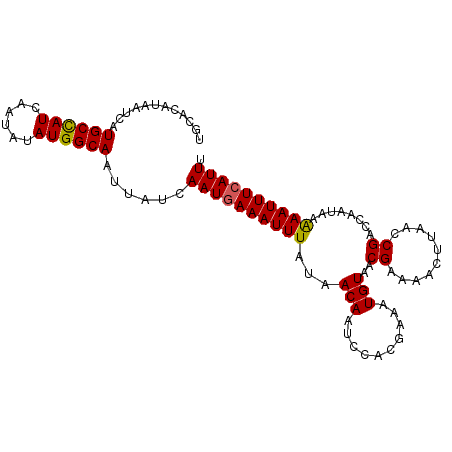
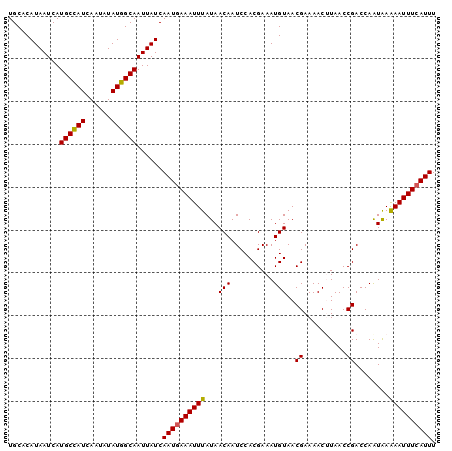
>2L_DroMel_CAF1 11956280 100 - 22407834 UGCACAUAAUCAUGCCAUCAAUAUAUGGCAAUUAUCAAUGAAAUUUAUAACAAUCCACGAAAUGUAACGAAAACUUAACCGACCAAUAAAAAUUUCAUUU ............((((((......))))))......((((((((((...(((..........)))..((..........))........)))))))))). ( -11.80) >DroVir_CAF1 159853 100 - 1 UGCACAUAAUCAUGCCAUCAAUAUAUGGCAAUUAUCAAUGAAAUUUAUAACAAUCCACGAAAUGUAACGAAAACAUAACCGACCAAUAAAAAUUUCAUUU ............((((((......))))))......((((((((((.((....((......((((.......))))....))....)).)))))))))). ( -12.40) >DroGri_CAF1 159736 100 - 1 UGCACAUAAUCAUGCUAUCAAUAUAUGGCAAUUAUCAAUGAAAUUUAUAACAAUCCACGAAAUGUAACGAAAACAUAACCGACCAAUAAAAAUUUCAUUU ............((((((......))))))......((((((((((.((....((......((((.......))))....))....)).)))))))))). ( -10.10) >DroSec_CAF1 175642 100 - 1 UGCACAUAAUCAUGCCAUCAAUAUAUGGCAAUUAUCAAUGAAAUUUAUAACAAUCCACGAAAUGUAACGAAAACUUAACCGACCAAUAAAAAUUUCAUUU ............((((((......))))))......((((((((((...(((..........)))..((..........))........)))))))))). ( -11.80) >DroSim_CAF1 142043 100 - 1 UGCACAUAAUCAUGCCAUCAAUAUAUGGCAAUUAUCAAUGAAAUUUAUAACAAUCCACGAAAUGUAACGAAAACUUAACCGACCAAUAAAAAUUUCAUUU ............((((((......))))))......((((((((((...(((..........)))..((..........))........)))))))))). ( -11.80) >DroEre_CAF1 150579 100 - 1 UGCACAUAAUCAUGCCAUCAAUAUAUGGCAAUUAUCAAUGAAAUUUAUAACAAUCCACGAAAUGUAACGAAAACUUAACCGACCAGUGAGAAUUUAAUUU ..(((.......((((((......))))))...........................((........))................)))............ ( -9.60) >consensus UGCACAUAAUCAUGCCAUCAAUAUAUGGCAAUUAUCAAUGAAAUUUAUAACAAUCCACGAAAUGUAACGAAAACUUAACCGACCAAUAAAAAUUUCAUUU ............((((((......))))))......((((((((((...(((..........)))..((..........))........)))))))))). (-10.84 = -10.73 + -0.11)

| Location | 11,956,318 – 11,956,409 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 92.36 |
| Mean single sequence MFE | -10.70 |
| Consensus MFE | -10.10 |
| Energy contribution | -10.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.24 |
| SVM RNA-class probability | 0.998817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
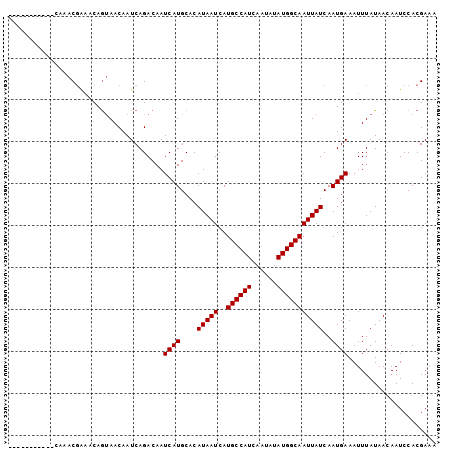
>2L_DroMel_CAF1 11956318 91 - 22407834 -----------CAAACGAAACAGUAACAAUCAGACAAUCAUGCACAUAAUCAUGCCAUCAAUAUAUGGCAAUUAUCAAUGAAAUUUAUAACAAUCCACGAAA -----------..........................((((....(((((..((((((......)))))))))))..))))..................... ( -10.10) >DroVir_CAF1 159891 88 - 1 --------------ACUGUGCAACAACAAUCAGACAAUCAUGCACAUAAUCAUGCCAUCAAUAUAUGGCAAUUAUCAAUGAAAUUUAUAACAAUCCACGAAA --------------..((((((..................))))))......((((((......))))))................................ ( -12.47) >DroSec_CAF1 175680 93 - 1 AA---------CAAACGAAACAGUAACAAUCAGACAAUCAUGCACAUAAUCAUGCCAUCAAUAUAUGGCAAUUAUCAAUGAAAUUUAUAACAAUCCACGAAA ..---------..........................((((....(((((..((((((......)))))))))))..))))..................... ( -10.10) >DroSim_CAF1 142081 91 - 1 -----------CAAACGAAACAGUAACAAUCAGACAAUCAUGCACAUAAUCAUGCCAUCAAUAUAUGGCAAUUAUCAAUGAAAUUUAUAACAAUCCACGAAA -----------..........................((((....(((((..((((((......)))))))))))..))))..................... ( -10.10) >DroEre_CAF1 150617 90 - 1 ------------AAACGAAACAGUAACAAUCAGACAAUCAUGCACAUAAUCAUGCCAUCAAUAUAUGGCAAUUAUCAAUGAAAUUUAUAACAAUCCACGAAA ------------.........................((((....(((((..((((((......)))))))))))..))))..................... ( -10.10) >DroYak_CAF1 146428 102 - 1 AAAAGCAAACGCAAACGAAACAGUAACAAUCAGACAAUCAUGCACAUAAUCAUGCCAUCAAUAUAUGGCAAUUAUCAAUGAAAUUUAUAACAAUCCACGAAA ....((....)).........................((((....(((((..((((((......)))))))))))..))))..................... ( -11.30) >consensus ___________CAAACGAAACAGUAACAAUCAGACAAUCAUGCACAUAAUCAUGCCAUCAAUAUAUGGCAAUUAUCAAUGAAAUUUAUAACAAUCCACGAAA .....................................((((....(((((..((((((......)))))))))))..))))..................... (-10.10 = -10.10 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:52 2006