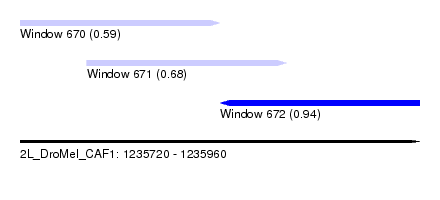
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,235,720 – 1,235,960 |
| Length | 240 |
| Max. P | 0.937335 |

| Location | 1,235,720 – 1,235,840 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -41.80 |
| Consensus MFE | -35.74 |
| Energy contribution | -36.80 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1235720 120 + 22407834 AUGCGUGCGGCAAAAGUUUUGCGGGCAGGAAGGCCAAAACUCGAGUGCAGCUCGCAGGACACGACCAGUGGAAUAAUCCUUUGUCCUGCCACGGAUGCUCUCCCCAAAAACUUUUGCGGU ......((.(((((((((((..(((..((....)).......(((.(((.((.(((((((((.....))(((....)))...)))))))...)).))).)))))).))))))))))).)) ( -43.50) >DroSec_CAF1 13050 120 + 1 AUGCGUGCGGCAAAAGUUUUACGGGCAGGAAGGCCAAAACUCGAGUGCAGCUCGCAGGACACGACCAGAGGAAUAAUCCUUUGUCCUGCCACGGAUGCUCUCCCCAAAAACUUUUGCGGU ......((.(((((((((((..(((..((....)).......(((.(((.((.((((((((......(((((....)))))))))))))...)).))).)))))).))))))))))).)) ( -44.40) >DroSim_CAF1 8833 120 + 1 AUGCGUGCGGCAAAAGUUUUGCGGGCAGGAAGGCCAAAACUCGAGUGCAGCUCGCAGGACACGACCAGUGGAAUAAUCCUUUGUCCUGCCACGGAUGCUCUCCCCAAAAACUUUUGCGGU ......((.(((((((((((..(((..((....)).......(((.(((.((.(((((((((.....))(((....)))...)))))))...)).))).)))))).))))))))))).)) ( -43.50) >DroYak_CAF1 9137 119 + 1 AUGCGUGCGGCAAAAGUUUUGUGGGCAGAAAGGCUAAGACUCGAGUGCAGCUCACAGGACACGACCAGUGGAAUAAUCCUUUGUCCUGCCACGGAUGUGCCUCCCAA-AAGUUUUGCGGU ......((.((((((.(((((.(((((.(..(((........(((.....)))..(((((((.....))(((....)))...)))))))).....).)))))..)))-)).)))))).)) ( -35.80) >consensus AUGCGUGCGGCAAAAGUUUUGCGGGCAGGAAGGCCAAAACUCGAGUGCAGCUCGCAGGACACGACCAGUGGAAUAAUCCUUUGUCCUGCCACGGAUGCUCUCCCCAAAAACUUUUGCGGU ......((.(((((((((((..(((.(((....((.......(((.....)))(((((((((.....).(((....)))..))))))))...))....))).))).))))))))))).)) (-35.74 = -36.80 + 1.06)

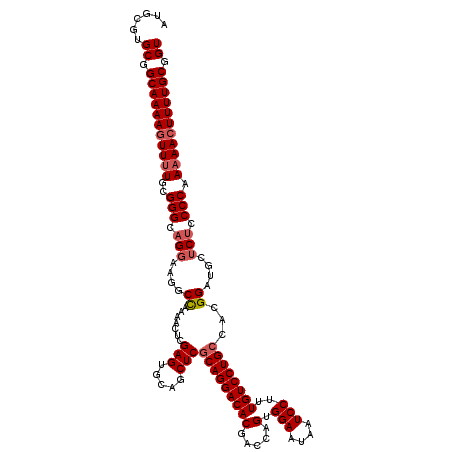
| Location | 1,235,760 – 1,235,880 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -34.67 |
| Consensus MFE | -29.92 |
| Energy contribution | -30.04 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1235760 120 + 22407834 UCGAGUGCAGCUCGCAGGACACGACCAGUGGAAUAAUCCUUUGUCCUGCCACGGAUGCUCUCCCCAAAAACUUUUGCGGUUCAAUUUGAAAAGGAAACUGCAAAGGAAAUUCCAAGGACA ..(((.(((.((.(((((((((.....))(((....)))...)))))))...)).))).)))........(((((((((((...(((....))).))))))))))).............. ( -35.00) >DroSec_CAF1 13090 120 + 1 UCGAGUGCAGCUCGCAGGACACGACCAGAGGAAUAAUCCUUUGUCCUGCCACGGAUGCUCUCCCCAAAAACUUUUGCGGUUCAAUUUGAAAAGGAAACUGCAAAGGAAAUUCCAAGGACA ..(((.(((.((.((((((((......(((((....)))))))))))))...)).))).)))........(((((((((((...(((....))).))))))))))).............. ( -37.50) >DroSim_CAF1 8873 120 + 1 UCGAGUGCAGCUCGCAGGACACGACCAGUGGAAUAAUCCUUUGUCCUGCCACGGAUGCUCUCCCCAAAAACUUUUGCGGUUCAAUUUGAAAAGGAAACUGCAAAGGAAAUUCCAAGGACA ..(((.(((.((.(((((((((.....))(((....)))...)))))))...)).))).)))........(((((((((((...(((....))).))))))))))).............. ( -35.00) >DroYak_CAF1 9177 119 + 1 UCGAGUGCAGCUCACAGGACACGACCAGUGGAAUAAUCCUUUGUCCUGCCACGGAUGUGCCUCCCAA-AAGUUUUGCGGUUCAAUUUGAAAAGAAAACUGCAAAGGAAAUUCCAAGGACA ..(((.....)))..(((.((((.((.((((.((((....))))....)))))).))))))).((..-....(((((((((...(((....))).)))))))))((.....))..))... ( -31.20) >consensus UCGAGUGCAGCUCGCAGGACACGACCAGUGGAAUAAUCCUUUGUCCUGCCACGGAUGCUCUCCCCAAAAACUUUUGCGGUUCAAUUUGAAAAGGAAACUGCAAAGGAAAUUCCAAGGACA ..(((.(((.((.(((((((((.....).(((....)))..))))))))...)).))).))).((.......(((((((((..............)))))))))((.....))..))... (-29.92 = -30.04 + 0.12)

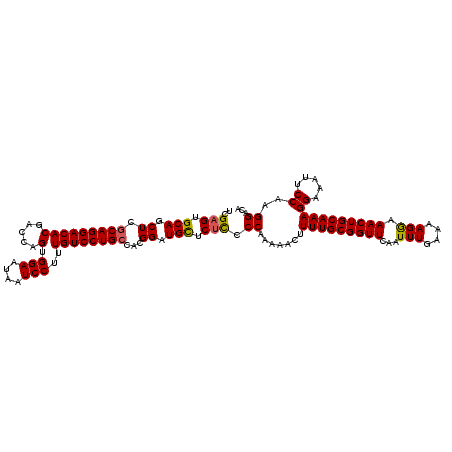
| Location | 1,235,840 – 1,235,960 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -28.12 |
| Consensus MFE | -25.13 |
| Energy contribution | -25.32 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1235840 120 - 22407834 UUCCUCUGAUAUCCCAGGGCCAUGUACAUUUUCAUUAGUUUAAUUAAAAUUUUAUUCAUCCUCGUGUGUGCGUGGCUGUGUGUCCUUGGAAUUUCCUUUGCAGUUUCCUUUUCAAAUUGA ((((...((((...((.((((((((((((........(..(((........)))..)........)))))))))))).))))))...)))).........((((((.......)))))). ( -27.99) >DroSec_CAF1 13170 120 - 1 UUCCUCUGAUAUCCCAGGGCCAUGUACAUUUUCAUUAGUUUAAUUAAAAUUUUAUUCAUCCUCGUGUGUGCGUGGCUGUGUGUCCUUGGAAUUUCCUUUGCAGUUUCCUUUUCAAAUUGA ((((...((((...((.((((((((((((........(..(((........)))..)........)))))))))))).))))))...)))).........((((((.......)))))). ( -27.99) >DroSim_CAF1 8953 120 - 1 UUCCUCUGAUAUCCCAGGGCCAUGUACAUUUUCAUUAGUUUAAUUAAAAUUUUAUUCAUCCUCGUGUGUGCGUGGCUGUGUGUCCUUGGAAUUUCCUUUGCAGUUUCCUUUUCAAAUUGA ((((...((((...((.((((((((((((........(..(((........)))..)........)))))))))))).))))))...)))).........((((((.......)))))). ( -27.99) >DroYak_CAF1 9256 120 - 1 UUCCUCUGGCAUCCAGGGGCCAUGUACAUAUUCAUUAGUUUAAUUAAAAUUGUAUUCAUCCUUGUGUGUGCGUGCCUGUGUGUCCUUGGAAUUUCCUUUGCAGUUUUCUUUUCAAAUUGA ((((...((((..((((.((((..((((.......((((((.....))))))..........))))..)).)).))))..))))...)))).........((((((.......)))))). ( -28.53) >consensus UUCCUCUGAUAUCCCAGGGCCAUGUACAUUUUCAUUAGUUUAAUUAAAAUUUUAUUCAUCCUCGUGUGUGCGUGGCUGUGUGUCCUUGGAAUUUCCUUUGCAGUUUCCUUUUCAAAUUGA ((((...((((...((.((((((((((((........(..(((........)))..)........)))))))))))).))))))...)))).........((((((.......)))))). (-25.13 = -25.32 + 0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:37 2006