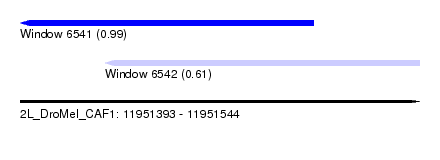
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,951,393 – 11,951,544 |
| Length | 151 |
| Max. P | 0.994551 |

| Location | 11,951,393 – 11,951,504 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.43 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -19.20 |
| Energy contribution | -20.24 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.994551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
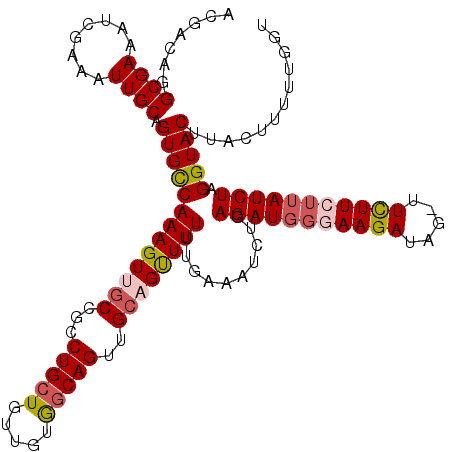
>2L_DroMel_CAF1 11951393 111 - 22407834 CUGCUGUUGUGGCAGUUGGAGUUUUUUGAAAUCUGAGAUGGGAAGAUAG-UUCUUCUUAUCUAGGUACUUACUUUUUGGUAGUACAGCUGCAAA--------UUUUAUAUCAUACACAUC (((((.....)))))(((.((((.........((.(((((((((((...-.)))))))))))))(((((.(((....)))))))))))).))).--------.................. ( -30.40) >DroSec_CAF1 170760 109 - 1 CUGCUGUUGUGGCAGUUGCAGCUUUUUGAAAUCUGAGAUGGGAAGAUAG-UUCUUCUAAUCUAGGUACAUACUUUUUGGUAGUACAGCUGCACU--------UUU--CAUUAUACACAUC (((((.....))))).(((((((.........((.((((.((((((...-.)))))).))))))((((.((((....)))))))))))))))..--------...--............. ( -32.90) >DroSim_CAF1 137187 109 - 1 CUGCUGUUGUGGCAGUUGCAGCUUUUUGAAAUCUGAGAUGGGAAGAUAG-UUCUUCUUAUCUAGGUACUUACUUUUUGGUAGUACAGCUGCACU--------UUU--UAUUACACACAUC (((((.....))))).(((((((.........((.(((((((((((...-.)))))))))))))(((((.(((....)))))))))))))))..--------...--............. ( -36.10) >DroEre_CAF1 145593 101 - 1 CUGCUGUUGCGCCAGUUGCUGUUUUCCGAAAACAGAGAUGCCAAGAUA--CUUUUCUUAUCUAGGUACUUACUU-UAAGUAGUACAAAUACACGGGA----------------AGAUAAC ..((((......))))...((((((((.......(((.((((.(((((--.......))))).)))))))....-...(((.......)))...)))----------------))))).. ( -20.30) >DroYak_CAF1 141100 113 - 1 CUGCUGUUGCAGCAGUUGCUGUUUUCCGAAAACAGAGAUGGCAAGAUAGAUUCUUCUUAUCUAGGUAC----UU-UGAAUAGUACUGAAAUACAGGAGCUUAGAA--UAUUGUAAGUAAC ((((((...))))))...((((((((.........((((((.((((.....)))).)))))).(((((----(.-.....)))))))))).))))..(((((.(.--...).)))))... ( -27.20) >consensus CUGCUGUUGUGGCAGUUGCAGUUUUUUGAAAUCUGAGAUGGGAAGAUAG_UUCUUCUUAUCUAGGUACUUACUUUUUGGUAGUACAGCUGCACA________UUU__UAUUAUACACAUC (((((.....))))).((.((((............(((((((((((.....)))))))))))..((((.((((....)))))))))))).))............................ (-19.20 = -20.24 + 1.04)



| Location | 11,951,425 – 11,951,544 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.77 |
| Mean single sequence MFE | -34.34 |
| Consensus MFE | -26.92 |
| Energy contribution | -27.80 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11951425 119 - 22407834 ACGACAGGCGAAAUCGAAAUUGCAGUGCCAAAGUUGCCGCCUGCUGUUGUGGCAGUUGGAGUUUUUUGAAAUCUGAGAUGGGAAGAUAG-UUCUUCUUAUCUAGGUACUUACUUUUUGGU ............(((((((....((((((((((...(((.(((((.....))))).)))...)))).........(((((((((((...-.))))))))))).))))))....))))))) ( -35.50) >DroSec_CAF1 170790 119 - 1 ACGACAGGCGAAAUCGAAAUUGCAGUGUCAAAGUUGCCGCCUGCUGUUGUGGCAGUUGCAGCUUUUUGAAAUCUGAGAUGGGAAGAUAG-UUCUUCUAAUCUAGGUACAUACUUUUUGGU ..((((.((((........))))..))))((((((((...(((((.....)))))..)))))))).....((((.((((.((((((...-.)))))).)))))))).............. ( -35.10) >DroSim_CAF1 137217 119 - 1 ACGACAGGCGAAAUCGAAAUUGCAGUGCCAAAGUUGCCGCCUGCUGUUGUGGCAGUUGCAGCUUUUUGAAAUCUGAGAUGGGAAGAUAG-UUCUUCUUAUCUAGGUACUUACUUUUUGGU ............(((((((....((((((((((((((...(((((.....)))))..))))))))..........(((((((((((...-.))))))))))).))))))....))))))) ( -40.60) >DroEre_CAF1 145617 117 - 1 ACGACAGGCGAAAUCGAAAUUGCAGUGCCAAAGUUGCCGCCUGCUGUUGCGCCAGUUGCUGUUUUCCGAAAACAGAGAUGCCAAGAUA--CUUUUCUUAUCUAGGUACUUACUU-UAAGU .((((.((((.......(((.((((.((..........)))))).))).)))).))))((((((.....))))))((.((((.(((((--.......))))).)))))).....-..... ( -27.70) >DroYak_CAF1 141138 115 - 1 AGGACAGGCGAAAUCGAAAUUGCAGUGCCAAAGUUGCCGCCUGCUGUUGCAGCAGUUGCUGUUUUCCGAAAACAGAGAUGGCAAGAUAGAUUCUUCUUAUCUAGGUAC----UU-UGAAU ....(((((((........))))((((((.....(((((.((.((((((((((....)))))........))))))).)))))(((((((....)).))))).)))))----))-))... ( -32.80) >consensus ACGACAGGCGAAAUCGAAAUUGCAGUGCCAAAGUUGCCGCCUGCUGUUGUGGCAGUUGCAGUUUUUUGAAAUCUGAGAUGGGAAGAUAG_UUCUUCUUAUCUAGGUACUUACUUUUUGGU .......((((........)))).(((((((((((((...(((((.....)))))..))))))))..........(((((((((((.....))))))))))).)))))............ (-26.92 = -27.80 + 0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:44 2006