| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,950,208 – 11,950,299 |
| Length | 91 |
| Max. P | 0.534582 |

| Location | 11,950,208 – 11,950,299 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 76.20 |
| Mean single sequence MFE | -26.63 |
| Consensus MFE | -15.55 |
| Energy contribution | -15.63 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534582 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11950208 91 + 22407834 AUCUA---------UGCCUGGCAUACAAAAUGCAUCUAAUUUUGGGCCUAACGACAACU-UGUCGCUUGUCAUGGCCU--CCUGGCCCGAGUCGCGUUGCUGC-- ...((---------(((...)))))......(((......((((((((....(((....-.)))(((......)))..--...))))))))......)))...-- ( -24.30) >DroVir_CAF1 154579 99 + 1 CUGUCUCUUUCGCAUG-CCGGCAUGU-AAAUGCCUCUAAUUUUGGACCUAACGACAACUUUGUCGUUUGUCAUGGCACAUGGAUGUCUU--UGGCAUUGUUGU-- ...............(-(((((((..-..)))))..........(((..(((((((....))))))).)))..)))(((..(((((...--..)))))..)))-- ( -29.00) >DroSec_CAF1 169472 91 + 1 AUCCA---------UGCCUGGCAUACAAAAUGCAUCUAAUUUUGGGCCUAACGACAACU-UGUCGCUUGUCAUGGCCU--CCUGGCCCGAGUCGCGUUGCUGC-- ....(---------(((...((((.....)))).......((((((((....(((....-.)))(((......)))..--...))))))))..))))......-- ( -24.10) >DroEre_CAF1 144153 93 + 1 AUCUA---------UGCCUGGCAUACAAAAUGCAUCUAAUUUUGGGCCUAACGACAACU-UGUCGCUUGUCAUGGCCU--CCUGGCCCGAGUCGCGUUGUUGUAG .....---------.....(((...((((((.......)))))).)))..((((((((.-.((.(((((....((((.--...))))))))).)))))))))).. ( -29.70) >DroYak_CAF1 139762 91 + 1 AUCUA---------UGCCUGGCAUACAAAAUGCAUCUAAUUUUGGGCCUAACGACAACU-UGUCGCUUGUCAUGGCCU--CCUGGCCCGAGUCGCGUUGUUGU-- .....---------.....(((...((((((.......)))))).)))..((((((((.-.((.(((((....((((.--...))))))))).))))))))))-- ( -29.00) >DroMoj_CAF1 162717 97 + 1 CUUUGUCUCUCGCUAG-CCGGCAUGU-AAAUGCCUCUAAUUUUGGGAAGAACGACAACU-UGUCUUUUGUCAUGGCCCAUGGAUGUCU---UGGCUUUGUAAU-- ...........((.((-(((((((..-..)))))((((....((((......(((((..-......)))))....)))))))).....---.))))..))...-- ( -23.70) >consensus AUCUA_________UGCCUGGCAUACAAAAUGCAUCUAAUUUUGGGCCUAACGACAACU_UGUCGCUUGUCAUGGCCU__CCUGGCCCGAGUCGCGUUGUUGU__ ..............(((...((((.....))))........(((((((....((((....))))(((......))).......)))))))...)))......... (-15.55 = -15.63 + 0.09)

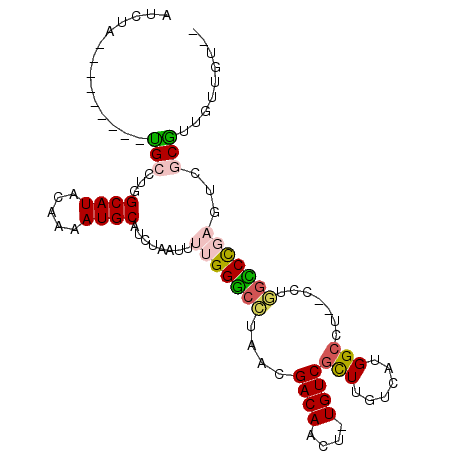

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:42 2006