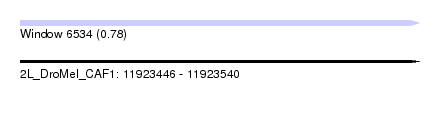
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,923,446 – 11,923,540 |
| Length | 94 |
| Max. P | 0.783841 |

| Location | 11,923,446 – 11,923,540 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 80.34 |
| Mean single sequence MFE | -21.40 |
| Consensus MFE | -11.34 |
| Energy contribution | -12.13 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.783841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
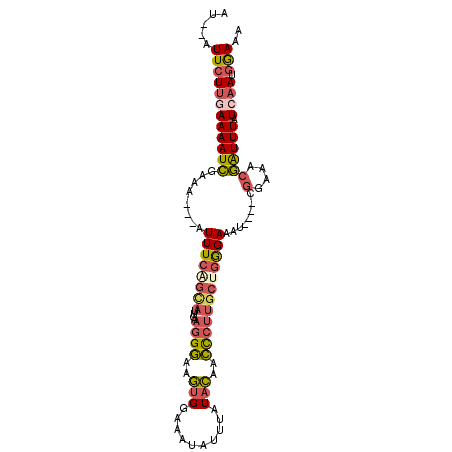
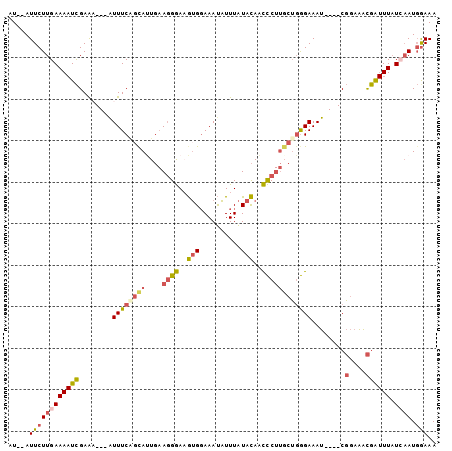
>2L_DroMel_CAF1 11923446 94 + 22407834 AU--AUUCUUGAAAAUCGAAA---AUUUCAGCAUUGAAGGGAAGUGGAAAUAUUUAUACAACCCUUUCUGGGAAAU----CGGAAACGAUUUAUCAAUGGAAA ..--.(((((((((.......---.))))))((((((((((..(((..........)))..)))).......((((----((....)))))).))))))))). ( -22.10) >DroSec_CAF1 142873 94 + 1 AU--AUUCUUGAAAAUCGAAA---AUUUCUGCAUUGAAGGGAAGUGGAAAUAUUUAUACAACCCUUGCUGGGAAAU----CGGAAACGAUUUAUAAAUGGAAA ..--.((((............---(((((..(...........)..)))))((((((....(((.....)))((((----((....)))))))))))))))). ( -20.50) >DroSim_CAF1 109423 94 + 1 AU--AUUCUUGAAAAUCGAAA---AUUUCAGCAUUGAAGGGAAGUGGAAAUAUUUAUACAACCCUUGCUGGGAAAU----CGGAAACGAUUUAUCAAUGGAAA ..--.(((((((.........---..(((((((....((((..(((..........)))..)))))))))))((((----((....)))))).)))).))).. ( -24.60) >DroEre_CAF1 114060 94 + 1 AU--AUUCUUGAAAAUCGAAU---GUUUCGUUAUUGAAGGGAUGUGGAAAUAUUUAUACAACCCUUGCUGGGAAAU----CGGAAACGAUUUAUCAAUGAAAA .(--((((.........))))---)(((((((....(((((.((((..........)))).)))))...(..((((----((....))))))..)))))))). ( -23.00) >DroWil_CAF1 167913 98 + 1 AUCAAUUCUUGAAA-UUGAGUGUCAUUUCAGCAAU--AUGAAAAAGCAAACUUUUGUAUAUUUC-AGCACAGAAAUAAAUCUUUGAUAG-UUUUGCAUCGAAA .((((((.....))-)))).(((..(((((.....--.)))))..)))..........((((((-......))))))....((((((.(-.....))))))). ( -13.10) >DroYak_CAF1 112077 94 + 1 AU--AUUCUUGAAAAUCGAAA---GUUUCAGCACUGAAGGGAUGUGGAAAUAUUUAUACAACCCUUGCUGGGAAAU----CGGAAGCGAUUUAUCAAUGGAAA ..--.(((((((((((((...---.(..(((((....((((.((((..........)))).)))))))))..)...----(....))))))).)))).))).. ( -25.10) >consensus AU__AUUCUUGAAAAUCGAAA___AUUUCAGCAUUGAAGGGAAGUGGAAAUAUUUAUACAACCCUUGCUGGGAAAU____CGGAAACGAUUUAUCAAUGGAAA .....((((((((((((........((((((((....((((..(((..........)))..))))))))))))........(....)))))).)))).))).. (-11.34 = -12.13 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:36 2006