
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,896,698 – 11,896,805 |
| Length | 107 |
| Max. P | 0.987566 |

| Location | 11,896,698 – 11,896,805 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 76.59 |
| Mean single sequence MFE | -26.73 |
| Consensus MFE | -14.51 |
| Energy contribution | -15.57 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.54 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11896698 107 - 22407834 UUAAUCCGU-GCCACAGGCGAACUAAUUUCUGUUUGCCUUUCAAUUUCCAAACGUUAAUUGCAACAAAUUGGCUUGGUCA--GGACCAGAGGUCUGACCUAUAACUAUUU- .........-((((.((((((((........))))))))..(((((..(....)..)))))........))))..(((((--(.(((...)))))))))...........- ( -29.60) >DroVir_CAF1 104576 85 - 1 ------CGUG---AUAAG-----UAAACUUUGUCAGGCUGUCAAUUUGUCGAGCUUAAUUGCAUCAAGUUGGCUUGGGUU--GGCAUA----CUUUAGG---GUCAUG--- ------((((---((...-----((((...((((((.(((((((((((....((......))..)))))))))..)).))--))))..----.))))..---))))))--- ( -24.80) >DroSec_CAF1 119495 107 - 1 UUAAUCCGU-GCCACAGGCGAACUAAUUUCUGUUUGCCUUUCAAUUUACAAAAGUUAAUUGCAACAAAUUGGCUUGGUCA--GGACCAGAGGUCUGUCCUAUAACUAUUU- .....(((.-((((.((((((((........))))))))..(((((.((....)).)))))........)))).)))..(--((((.((....)))))))..........- ( -28.80) >DroSim_CAF1 85330 107 - 1 UUAAUCCGU-GCCACAGGCGAACUAAUUUCUGUUUGCCUUUCAAUUUCCAAACGUUAAUUGCAACAAAUUGGCUUGGUCA--GGACCAGAGGUCUGUCCUAUAACUAUUU- .....(((.-((((.((((((((........))))))))..(((((..(....)..)))))........)))).)))..(--((((.((....)))))))..........- ( -28.40) >DroEre_CAF1 90846 105 - 1 UUAAUCCGU-GCCACAAGCGAACUAAUUUCUGUUUGCUUUCCAAUUUCCAAACGUUAAUUGCAACAAAUUGGCUUGGCCA--GUACCAG--GUCUGUCCUAUAACUAUUU- .......(.-((((.((((((((........))))))))..............(((((((......))))))).)))))(--((...((--(.....)))...)))....- ( -21.50) >DroYak_CAF1 87430 108 - 1 UUAAUCCGU-GCCACAGGCGAACUAAUUUCUGUUUGCCUUUCAAUUUCCCAACGUUAAUUGCAACAAAUUGGCUUGGUCAAAGGACCAA--ACUCGCACCAUAACUAUUUU .......((-((...((((((((........))))))))..............(((((((......)))))))((((((....))))))--....))))............ ( -27.30) >consensus UUAAUCCGU_GCCACAGGCGAACUAAUUUCUGUUUGCCUUUCAAUUUCCAAACGUUAAUUGCAACAAAUUGGCUUGGUCA__GGACCAG__GUCUGUCCUAUAACUAUUU_ ...............((((((((........))))))))..............(((((((......)))))))((((((....))))))...................... (-14.51 = -15.57 + 1.06)

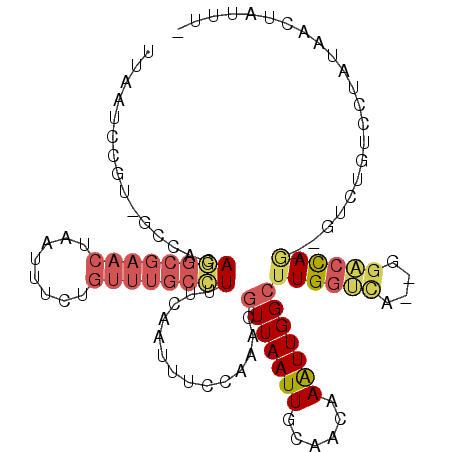
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:25 2006