
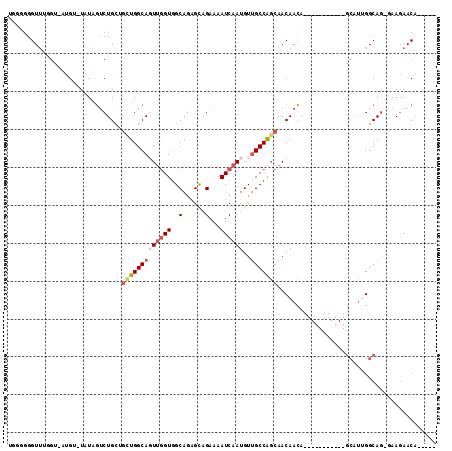
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,892,965 – 11,893,088 |
| Length | 123 |
| Max. P | 0.898677 |

| Location | 11,892,965 – 11,893,059 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 74.15 |
| Mean single sequence MFE | -27.55 |
| Consensus MFE | -12.16 |
| Energy contribution | -12.97 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.44 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11892965 94 + 22407834 UAGGGGGUUUGGU-AUGU-UAUAGUCUGCUGCUGGCAGUUGGUGGCAGAGCAGAAAAUCAAUGUUGCCAGCAACAACA-----------GCAUUGGCAG-GAAGAACA----- .............-.(((-(....((((((((((...((((.(((((..(((.........)))))))).))))..))-----------))...)))))-)...))))----- ( -29.60) >DroVir_CAF1 100736 87 + 1 -AAUGUGU------GCGAUUGUGCGCUGUGGGUGGCAUUUAGUAGCAGAGCUGAAAAUCAAUGUAGCCGCCAACAAGA--------------AGGGCAAAUGAGAACA----- -..(((..------.((.((((.(..(((.((((((.((((((......)))))).((....)).)))))).)))...--------------.).)))).))...)))----- ( -21.40) >DroSec_CAF1 115762 91 + 1 UGGGGGGUUUGGU-AUGU-UAUAGUCUG---CUGGCAGUUGGUGGCAGAGUAGAAAAUCAAUGUUGCCAGCAACAACA-----------GCAUUGGCAG-GAAGAACA----- ......((((...-.(((-((.....((---(((...((((.((((((.((.(.....).)).)))))).))))..))-----------))).))))).-...)))).----- ( -27.30) >DroSim_CAF1 81267 94 + 1 UGGGGGGUUUGGU-AUGU-UAUAGUCUGCUGCUGGCAGUUGGUGGCAGAGUAGAAAAUCAAUGUUGCCAGCAACAACA-----------GCAUUGGCAG-GAAGAACA----- .............-.(((-(....((((((((((...((((.((((((.((.(.....).)).)))))).))))..))-----------))...)))))-)...))))----- ( -29.80) >DroYak_CAF1 83629 100 + 1 UGUGGGGUUGGGUUUUGG-UAUAGUCUGCAGGUGGCAGUUGGUGGCAGGGCAGAAAAUCAAUGUUGCCAGCAACAACA-----------GCAUGGGCAG-GAAGAACAGGGAG ((((..(((((...((((-(....(((((..((.((.....)).))...)))))..))))).....)))))..).)))-----------((....))..-............. ( -24.50) >DroAna_CAF1 96959 104 + 1 UU-AAGGUUCAGG-AGGC-UAUAGUCUGCUGCUGGCAUUCGGUGGCAAAGCAGAAAAUCAAUGUUGCCAGCAACAACAUUGCCAGCGCGGCAUUGGCAG-AAACAACC----- ..-..((((....-..((-((..(.((((.(((((((...(.((((((....(.....)....)))))).)........))))))))))))..))))..-....))))----- ( -32.70) >consensus UGGGGGGUUUGGU_AUGU_UAUAGUCUGCUGCUGGCAGUUGGUGGCAGAGCAGAAAAUCAAUGUUGCCAGCAACAACA___________GCAUUGGCAG_GAAGAACA_____ ..............................(((((((((((((..(......)...))))))..))))))).......................................... (-12.16 = -12.97 + 0.81)


| Location | 11,892,965 – 11,893,059 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 74.15 |
| Mean single sequence MFE | -20.53 |
| Consensus MFE | -10.49 |
| Energy contribution | -10.71 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11892965 94 - 22407834 -----UGUUCUUC-CUGCCAAUGC-----------UGUUGUUGCUGGCAACAUUGAUUUUCUGCUCUGCCACCAACUGCCAGCAGCAGACUAUA-ACAU-ACCAAACCCCCUA -----((((....-((((...(((-----------((..((((.(((((.....(.....).....))))).))))...))))))))).....)-))).-............. ( -22.70) >DroVir_CAF1 100736 87 - 1 -----UGUUCUCAUUUGCCCU--------------UCUUGUUGGCGGCUACAUUGAUUUUCAGCUCUGCUACUAAAUGCCACCCACAGCGCACAAUCGC------ACACAUU- -----................--------------...(((.((.(((......(.....)(((...))).......))).)).)))(((......)))------.......- ( -12.00) >DroSec_CAF1 115762 91 - 1 -----UGUUCUUC-CUGCCAAUGC-----------UGUUGUUGCUGGCAACAUUGAUUUUCUACUCUGCCACCAACUGCCAG---CAGACUAUA-ACAU-ACCAAACCCCCCA -----........-.......(((-----------((..((((.(((((.....(.....).....))))).))))...)))---)).......-....-............. ( -17.70) >DroSim_CAF1 81267 94 - 1 -----UGUUCUUC-CUGCCAAUGC-----------UGUUGUUGCUGGCAACAUUGAUUUUCUACUCUGCCACCAACUGCCAGCAGCAGACUAUA-ACAU-ACCAAACCCCCCA -----((((....-((((...(((-----------((..((((.(((((.....(.....).....))))).))))...))))))))).....)-))).-............. ( -22.70) >DroYak_CAF1 83629 100 - 1 CUCCCUGUUCUUC-CUGCCCAUGC-----------UGUUGUUGCUGGCAACAUUGAUUUUCUGCCCUGCCACCAACUGCCACCUGCAGACUAUA-CCAAAACCCAACCCCACA .............-((((....((-----------.((((..((.((((............))))..))...)))).)).....))))......-.................. ( -16.70) >DroAna_CAF1 96959 104 - 1 -----GGUUGUUU-CUGCCAAUGCCGCGCUGGCAAUGUUGUUGCUGGCAACAUUGAUUUUCUGCUUUGCCACCGAAUGCCAGCAGCAGACUAUA-GCCU-CCUGAACCUU-AA -----((((((.(-(((((......).(((((((......(((.((((((....(.....)....)))))).))).))))))).)))))..)))-))).-..........-.. ( -31.40) >consensus _____UGUUCUUC_CUGCCAAUGC___________UGUUGUUGCUGGCAACAUUGAUUUUCUGCUCUGCCACCAACUGCCAGCAGCAGACUAUA_ACAU_ACCAAACCCCCCA .....................................((((.(((((((...(((.................))).))))))).))))......................... (-10.49 = -10.71 + 0.22)


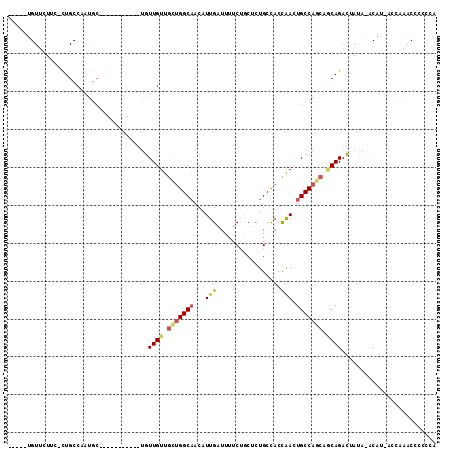
| Location | 11,892,998 – 11,893,088 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 97.04 |
| Mean single sequence MFE | -22.17 |
| Consensus MFE | -19.40 |
| Energy contribution | -19.73 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11892998 90 + 22407834 CAGUUGGUGGCAGAGCAGAAAAUCAAUGUUGCCAGCAACAACAGCAUUGGCAGGAAGAACAGUAGGAGCUGGAGGAACUGGAGGAGCAGG ..((((.(((((..(((.........)))))))).))))....((.((..(((......((((....))))......)))..)).))... ( -23.80) >DroSec_CAF1 115792 90 + 1 CAGUUGGUGGCAGAGUAGAAAAUCAAUGUUGCCAGCAACAACAGCAUUGGCAGGAAGAACAGGAGGAGCUGGAGGAACUGGAGGAACAGG ..((((.((((((.((.(.....).)).)))))).))))..((((.((..(..(.....)..)..))))))......(((......))). ( -22.80) >DroSim_CAF1 81300 90 + 1 CAGUUGGUGGCAGAGUAGAAAAUCAAUGUUGCCAGCAACAACAGCAUUGGCAGGAAGAACAGGAGGAGCUGGAGGAAGUGGAGGAACAGG ..((((.((((((.((.(.....).)).)))))).))))..((((.((..(..(.....)..)..))))))......((......))... ( -19.90) >consensus CAGUUGGUGGCAGAGUAGAAAAUCAAUGUUGCCAGCAACAACAGCAUUGGCAGGAAGAACAGGAGGAGCUGGAGGAACUGGAGGAACAGG ..((((.((((((....(.....)....)))))).))))..((((.((..(.(......).)..)).))))......(((......))). (-19.40 = -19.73 + 0.33)

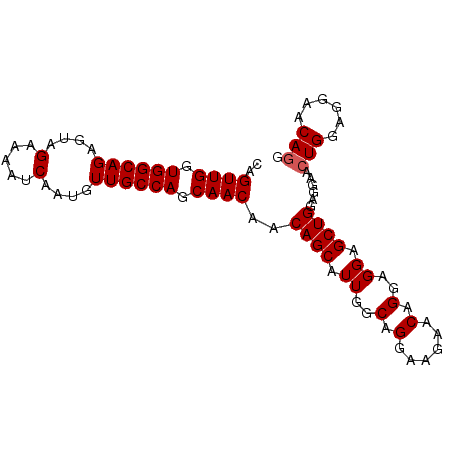
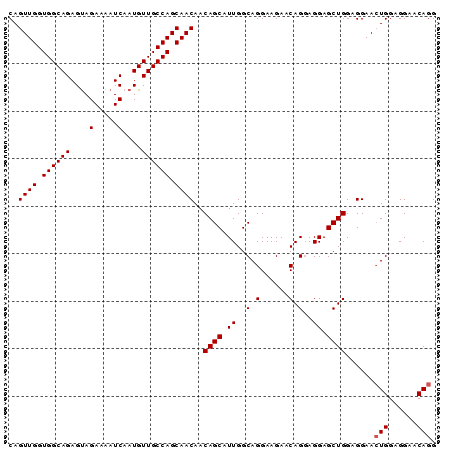
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:24 2006