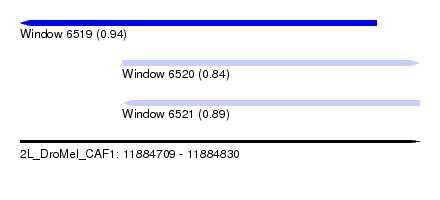
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,884,709 – 11,884,830 |
| Length | 121 |
| Max. P | 0.937240 |

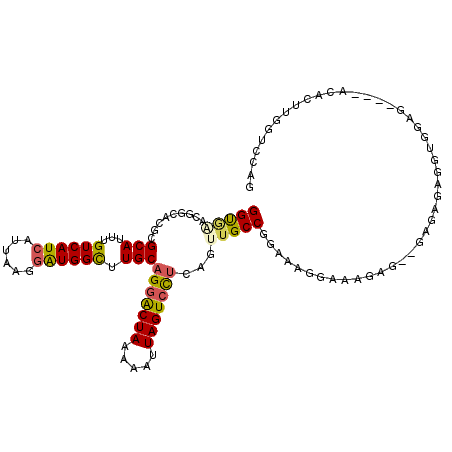
| Location | 11,884,709 – 11,884,817 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 76.53 |
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -19.02 |
| Energy contribution | -19.05 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937240 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
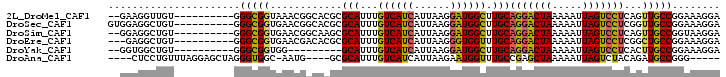
>2L_DroMel_CAF1 11884709 108 - 22407834 GGUAAACGGCACGCGCAUUUGUCAUCAUUAAGGAUGGCUUGCAGGACUAAAAAUUAGUCCUCAGUUGCCGGAAAGGAAAGAG--GAGAGAGGUGGAG----ACACUUGGUCCAG ......(((((.(((((...((((((......)))))).)))(((((((.....)))))))..)))))))...........(--((...(((((...----.)))))..))).. ( -39.40) >DroSec_CAF1 107486 108 - 1 GGUGAACGGCACGCGCAUUUGUCAUCAUUAAGGAUGGCUUGCAGGACUAAAAAUUAGUCCUCGGUUGCCGGAAAGGAAAGAG--GAGAGAGGUGGCG----ACACUUGGUCCAG ......(((((.(((((...((((((......)))))).)))(((((((.....)))))))..)))))))...........(--((...(((((...----.)))))..))).. ( -37.70) >DroSim_CAF1 73053 108 - 1 GGUGAACGGCAAGCGCAUUUGUCAUCAUUAAGGAUGGCUUGCAGGACUAAAAAUUAGUCCUCAGUUGCCGGUAAGGAAAGAG--GAGAGAGGUGGCG----ACAAUUGGUCCAG ......((((((..(((...((((((......)))))).)))(((((((.....)))))))...))))))...........(--((..((..((...----.)).))..))).. ( -35.30) >DroEre_CAF1 78841 111 - 1 GGUGAACGACACGCGCAUUUGUCAUCAUUAAGGGUGGUUUGCAGGACUAAAAAUUAGUCCUCGGCUGCCGGAAAGGAAAGUG--GAGACCG-GGCAGGGGUAUCCUUGGUCCAG (((...(..((((((((....(((((......)))))..)))(((((((.....)))))))..))..((.....))...)))--..))))(-(((((((....)))).)))).. ( -33.50) >DroYak_CAF1 75353 101 - 1 GGUGG---------GCAUUUGUCAUCAUUAAGGAUGGCUUGCAGGACUAAAAAUUAGUCCUCACUUGCCGGAAAGGAGAGAGAGGAGAGAGGGGCAG----ACCCUUGGUCCAG .....---------(((...((((((......)))))).))).(((((.........(((((.((..((.....))..)).)))))..(((((....----.)))))))))).. ( -38.40) >DroAna_CAF1 82492 83 - 1 GGC-AAUG----GCGCAUUUGUCAUCAUUAAGAAUGGUUUGCCGAGCUAAAAAUUAGUCUACAGAUGCCGGG----------------------GCC----CGUUAUGUUCCAC (((-..((----((..((((((....(((((...((((((...))))))....)))))..))))))))))..----------------------)))----............. ( -17.90) >consensus GGUGAACGGCACGCGCAUUUGUCAUCAUUAAGGAUGGCUUGCAGGACUAAAAAUUAGUCCUCAGUUGCCGGAAAGGAAAGAG__GAGAGAGGUGGAG____ACACUUGGUCCAG (((((.........(((...((((((......)))))).)))(((((((.....)))))))...)))))............................................. (-19.02 = -19.05 + 0.03)

| Location | 11,884,740 – 11,884,830 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 79.29 |
| Mean single sequence MFE | -20.57 |
| Consensus MFE | -14.14 |
| Energy contribution | -14.25 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.839021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11884740 90 + 22407834 UCCUUUCCGGCAACUGAGGACUAAUUUUUAGUCCUGCAAGCCAUCCUUAAUGAUGACAAAUGCGCGUGCCGUUUACCGCCC----------ACAACCUUC-- .......(((((....(((((((.....)))))))(((.(.((((......)))).)...)))...)))))..........----------.........-- ( -21.10) >DroSec_CAF1 107517 92 + 1 UCCUUUCCGGCAACCGAGGACUAAUUUUUAGUCCUGCAAGCCAUCCUUAAUGAUGACAAAUGCGCGUGCCGUUCACCGCCC----------ACAGCCUCCAC ........(((.....(((((((.....)))))))....(.((((......)))).)....(((.(((.....))))))..----------...)))..... ( -21.60) >DroSim_CAF1 73084 90 + 1 UCCUUACCGGCAACUGAGGACUAAUUUUUAGUCCUGCAAGCCAUCCUUAAUGAUGACAAAUGCGCUUGCCGUUCACCGCCC----------ACAGCCUCC-- ........(((.....(((((((.....)))))))((((((((((......))))........))))))............----------...)))...-- ( -23.20) >DroEre_CAF1 78875 89 + 1 UCCUUUCCGGCAGCCGAGGACUAAUUUUUAGUCCUGCAAACCACCCUUAAUGAUGACAAAUGCGCGUGUCGUUCACCGCCC----------ACAGCCUC--- ........(((.((..(((((((.....)))))))))........................(((.(((.....))))))..----------...)))..--- ( -21.90) >DroYak_CAF1 75386 81 + 1 UCCUUUCCGGCAAGUGAGGACUAAUUUUUAGUCCUGCAAGCCAUCCUUAAUGAUGACAAAUGC---------CCACCGCCC----------ACAGCCACC-- ........(((..((((((((((.....)))))))(((.(.((((......)))).)...)))---------........)----------)).)))...-- ( -19.60) >DroAna_CAF1 82508 88 + 1 -----CCCGGCAUCUGUAGACUAAUUUUUAGCUCGGCAAACCAUUCUUAAUGAUGACAAAUGCGC----CAUU-GCCACCCUAGCUCCUAAACAGGAG---- -----...((((......(((((.....))).))(((.......((........)).......))----)..)-))).......(((((....)))))---- ( -16.04) >consensus UCCUUUCCGGCAACUGAGGACUAAUUUUUAGUCCUGCAAGCCAUCCUUAAUGAUGACAAAUGCGCGUGCCGUUCACCGCCC__________ACAGCCUCC__ ........(((.....(((((((.....)))))))(((...((((......)))).....)))..............)))...................... (-14.14 = -14.25 + 0.11)

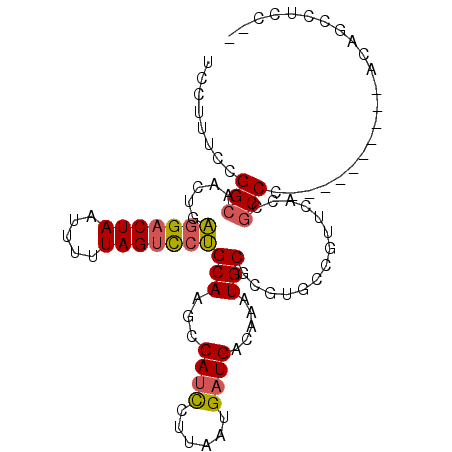

| Location | 11,884,740 – 11,884,830 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 79.29 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -21.25 |
| Energy contribution | -21.03 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
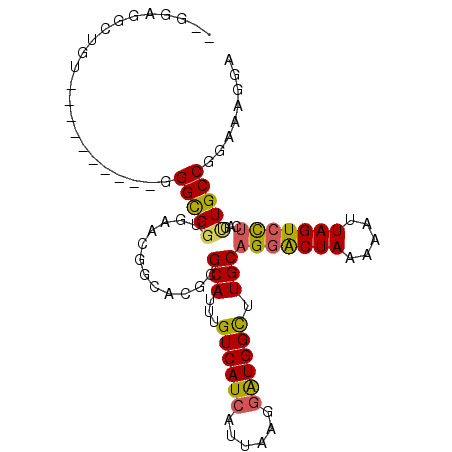
>2L_DroMel_CAF1 11884740 90 - 22407834 --GAAGGUUGU----------GGGCGGUAAACGGCACGCGCAUUUGUCAUCAUUAAGGAUGGCUUGCAGGACUAAAAAUUAGUCCUCAGUUGCCGGAAAGGA --.........----------..........(((((.(((((...((((((......)))))).)))(((((((.....)))))))..)))))))....... ( -28.70) >DroSec_CAF1 107517 92 - 1 GUGGAGGCUGU----------GGGCGGUGAACGGCACGCGCAUUUGUCAUCAUUAAGGAUGGCUUGCAGGACUAAAAAUUAGUCCUCGGUUGCCGGAAAGGA .(((..(((((----------(.((.(....).)).)))(((...((((((......)))))).)))(((((((.....))))))).)))..)))....... ( -32.50) >DroSim_CAF1 73084 90 - 1 --GGAGGCUGU----------GGGCGGUGAACGGCAAGCGCAUUUGUCAUCAUUAAGGAUGGCUUGCAGGACUAAAAAUUAGUCCUCAGUUGCCGGUAAGGA --....((((.----------...))))...((((((..(((...((((((......)))))).)))(((((((.....)))))))...))))))....... ( -32.10) >DroEre_CAF1 78875 89 - 1 ---GAGGCUGU----------GGGCGGUGAACGACACGCGCAUUUGUCAUCAUUAAGGGUGGUUUGCAGGACUAAAAAUUAGUCCUCGGCUGCCGGAAAGGA ---(((((.((----------(.(((((.....)).))).)))..))).))......((..(((.(.(((((((.....)))))))))))..))........ ( -28.90) >DroYak_CAF1 75386 81 - 1 --GGUGGCUGU----------GGGCGGUGG---------GCAUUUGUCAUCAUUAAGGAUGGCUUGCAGGACUAAAAAUUAGUCCUCACUUGCCGGAAAGGA --.....((.(----------.(((((((.---------(((...((((((......)))))).)))(((((((.....)))))))))).)))).)...)). ( -28.80) >DroAna_CAF1 82508 88 - 1 ----CUCCUGUUUAGGAGCUAGGGUGGC-AAUG----GCGCAUUUGUCAUCAUUAAGAAUGGUUUGCCGAGCUAAAAAUUAGUCUACAGAUGCCGGG----- ----(((((....)))))....((((((-((..----......))))))))........((((......(((((.....))).))......))))..----- ( -19.40) >consensus __GGAGGCUGU__________GGGCGGUGAACGGCACGCGCAUUUGUCAUCAUUAAGGAUGGCUUGCAGGACUAAAAAUUAGUCCUCAGUUGCCGGAAAGGA ......................(((((............(((...((((((......)))))).)))(((((((.....)))))))...)))))........ (-21.25 = -21.03 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:21 2006