| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,843,735 – 11,843,842 |
| Length | 107 |
| Max. P | 0.549990 |

| Location | 11,843,735 – 11,843,842 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.44 |
| Mean single sequence MFE | -31.43 |
| Consensus MFE | -14.95 |
| Energy contribution | -15.65 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
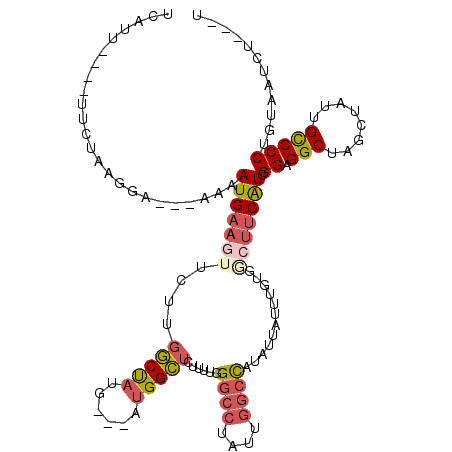
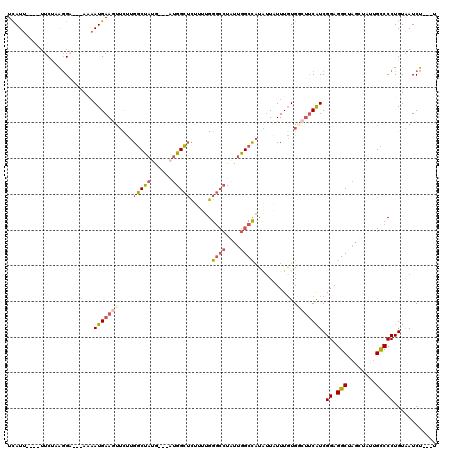
>2L_DroMel_CAF1 11843735 107 + 22407834 UCAUU----UUCUGAGGA---AAAAUGAAGUUCUUGGCUAUG---AUGGCUCUUUUGGGCCUAUUGGCUAUAUUCUUUGUGUCUUCAUCGGAGGCUAGUUAUUGCCCCUGUAAUCU---U .....----...((((((---...(..(((....((((((..---..(((((....)))))...))))))....)))..).))))))..((.(((........)))))........---. ( -33.50) >DroSec_CAF1 67072 107 + 1 UCAUU----UUCCAAGGA---AUAAUGAAGUUCUUGGCUAUA---AUGGCUUUUUUGGGCCUAUUGGCUAUAUUGUUUGUGGAUUCAUCGGAGGCUAGUUAUUGCCCCUGUAAUCU---U .....----.((((..((---((((((.......((((((..---..(((((....)))))...)))))))))))))).))))......((.(((........)))))........---. ( -27.31) >DroSim_CAF1 32464 107 + 1 UCAUU----UUCUAAGGA---AAAAUGAAGUUCUUGGCUAUG---AUGGCUCUUUUGGGGCUAUUGGCUAUAUUAUUUCUGGCUUCAUCGGAGGCUAGUUAUUGCCCCUGUAAUCU---U .....----.....(((.---..(((((......(((((..(---(((((((.....)))))))))))))........((((((((....)))))))))))))...))).......---. ( -28.50) >DroEre_CAF1 32885 107 + 1 UCAUU----UUCUUGCGC---AAAAUGAAGUUCUUGGCCAUG---CUGGCUCUUUUGGGCCUAUUGGCCAUGUUAUUUGUGGCUCCAUCGGAGGCAAGCUAUUGUCCCUGUAAUCU---U .....----...((((((---...(..((...(.((((((..---..(((((....)))))...)))))).)...))..).))......((.(((((....))))))).))))...---. ( -32.50) >DroYak_CAF1 33550 107 + 1 UCAUU----UUCUAGGGC---AAUAUGAAUUUCUUGGCUAUG---AUGGCGCUUUUGGGCCUUUUGGCCAUGUUAUUUGUGCCAUCAUCGGAGGCCAGCUACUGUCCCUGUAAUCU---C .....----...((((((---(....(.....)(((((((((---(((((((.....((((....)))).........))))))))))....))))))....)).)))))......---. ( -34.84) >DroAna_CAF1 39501 120 + 1 UUAUUAUCAUUCGGAGGAGCGACAAUGAAGUGUCUGACUUUGGUGCUGGUUCUAUUGGCCAUGUUGGCCUCCCUGUUUGUGACUUCGUCGGAGGCCUCCUACUGUCCCUGUGAUCUGUCC .....(((((.(((((((((((((....((..((.......))..))((((.....)))).)))))(((((((.....).(((...)))))))))))))).))).....)))))...... ( -31.90) >consensus UCAUU____UUCUAAGGA___AAAAUGAAGUUCUUGGCUAUG___AUGGCUCUUUUGGGCCUAUUGGCCAUAUUAUUUGUGGCUUCAUCGGAGGCUAGCUAUUGCCCCUGUAAUCU___U ........................(((((((....(((((......)))))......((((....))))............))))))).((.(((........)))))............ (-14.95 = -15.65 + 0.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:13 2006