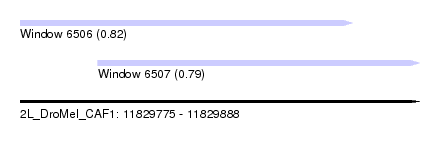
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,829,775 – 11,829,888 |
| Length | 113 |
| Max. P | 0.819059 |

| Location | 11,829,775 – 11,829,869 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 90.20 |
| Mean single sequence MFE | -21.30 |
| Consensus MFE | -19.32 |
| Energy contribution | -19.10 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.819059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
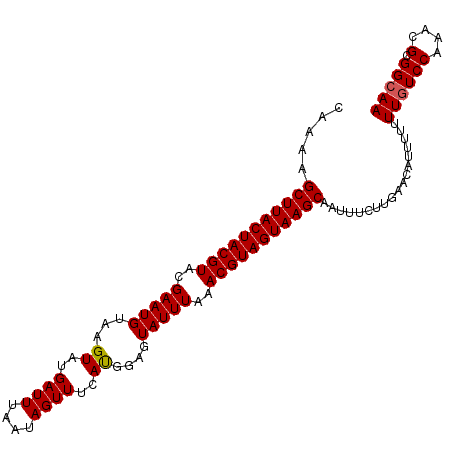
>2L_DroMel_CAF1 11829775 94 + 22407834 UAUCUUCCAGUUCUCACUCCUUCAAGUGCUUACUACGUAUGAAUGUAAGUUUGAUUUAAUAGUUGCACGGAGUAUUUAAACGUAGUAAGCAAUU-------- ..........................((((((((((((.((((((...((.(((((....))))).))....)))))).))))))))))))...-------- ( -22.70) >DroSec_CAF1 52996 102 + 1 UAUCUUCCAGUUUUCACUCCUUCAAAAGCUUACUACGUACGAAUGUAAGUAUGAUUUAAUAGUUUCAUGGAGUAUUUAAACGUAGUAAGCAAUUUCUUGAAC ...........................(((((((((((..(((((....(((((..........)))))...)))))..)))))))))))............ ( -20.60) >DroSim_CAF1 18550 102 + 1 UAUCUUCCAGUUCUCACUCCUUCAAAAGCUUACUACGUACGAAUGUAAGUAUGAUUUAAUAGUUUCAUGGAGUAUUUAAACGUAGUAAGCAAUUUCUUGAAC ...........................(((((((((((..(((((....(((((..........)))))...)))))..)))))))))))............ ( -20.60) >consensus UAUCUUCCAGUUCUCACUCCUUCAAAAGCUUACUACGUACGAAUGUAAGUAUGAUUUAAUAGUUUCAUGGAGUAUUUAAACGUAGUAAGCAAUUUCUUGAAC ...........................(((((((((((..(((((...((..((((....))))..))....)))))..)))))))))))............ (-19.32 = -19.10 + -0.22)


| Location | 11,829,797 – 11,829,888 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 87.06 |
| Mean single sequence MFE | -22.77 |
| Consensus MFE | -19.55 |
| Energy contribution | -19.67 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11829797 91 + 22407834 CAAGUGCUUACUACGUAUGAAUGUAAGUUUGAUUUAAUAGUUGCACGGAGUAUUUAAACGUAGUAAGCAAUU------------UUUUGUCCAAACGCGGCAA ....((((((((((((.((((((...((.(((((....))))).))....)))))).))))))))))))...------------..((((((....).))))) ( -24.80) >DroSec_CAF1 53018 103 + 1 CAAAAGCUUACUACGUACGAAUGUAAGUAUGAUUUAAUAGUUUCAUGGAGUAUUUAAACGUAGUAAGCAAUUUCUUGAACAUUUUUUUGUCCAAACGCGGCAA .....(((((((((((..(((((....(((((..........)))))...)))))..)))))))))))......(((.(((......))).)))......... ( -22.90) >DroSim_CAF1 18572 103 + 1 CAAAAGCUUACUACGUACGAAUGUAAGUAUGAUUUAAUAGUUUCAUGGAGUAUUUAAACGUAGUAAGCAAUUUCUUGAACAUUUUUUUUUCCAUACGCGGCAA .....(((((((((((..(((((....(((((..........)))))...)))))..)))))))))))................................... ( -20.60) >consensus CAAAAGCUUACUACGUACGAAUGUAAGUAUGAUUUAAUAGUUUCAUGGAGUAUUUAAACGUAGUAAGCAAUUUCUUGAACAUUUUUUUGUCCAAACGCGGCAA .....(((((((((((..(((((...((..((((....))))..))....)))))..)))))))))))..................((((((....).))))) (-19.55 = -19.67 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:07 2006